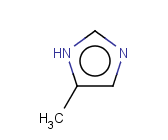
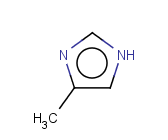
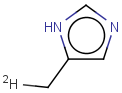
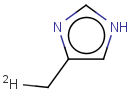
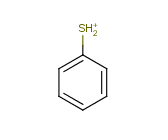
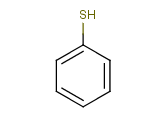
Tautomer search / Vague bond search / sp-Hybridization
A few selected search options are described below.
Tautomer search option
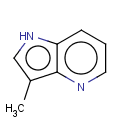
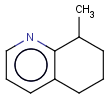
This search option can instruct the search engine to look for all tautomer forms of the query, as generated by the Isomers/Tautomers plugin in Marvin. (For alternative solutions to handle tautomers, see JChem Database Concepts.)
The following options are available in tautomer search:
-
tautomer search on:
tautomers of the query and the target are taken into account; -
tautomer search on with ignore stereo information in tautomer regions:
tautomers of the query and the target are taken into account;
double bond stereo information and tetrahedral stereo information of the query and the target structures in the tautomer regions are not considered during the search;
avaliable for duplicate, full structure, and full fragment searches; -
tautomer search off:
tautomers of the query and the target are not taken into account.
You can find information about how to use tautomer search options on different JChem platforms here.
Duplicate, full and full fragment searches are performed using the generic tautomer form of the query and the target in non-markush tables and in memory which makes the search very effective.
Remark: duplicate search in a table created with "Duplicate search uses tautomers" option results in a tautomer duplicate search except when "Tautomer search" option is explicitly switched off.
The following restrictions apply in tautomer search mode:
-
The query must not have any query features, and
-
This search option is best suited to full or full fragment search, as the tautomers of the query are generated for a whole molecule and not for a substructure.
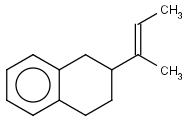
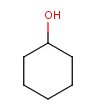
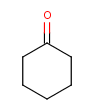
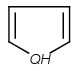
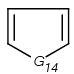
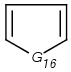
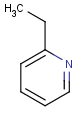
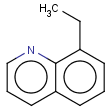
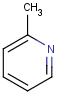
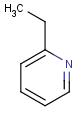
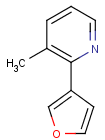
Table 1. Tautomer search examples
|
Query |
Target |
|||
|
Tautomer searching off |
Tautomer searching on |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table 2.
Examples for duplicate search with 'Tautomer search with ignore stereo information in tautomer region' option
Note: Because the symmetric nature of duplicate search, the roles of query and target molecules are exchangable.
|
Query |
Target |
Tautomer searching off |
Tautomer searching on |
Tautomer searching on with ignore stereo information in tautomer region |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Tautomer duplicate filtering
JChem tables can be created with the setting "Duplicate search uses tautomers". See Administration guide of JChem. Duplicate search in these tables will consider tautomers as duplicates by default. It can be switched off by the tautomerSearch:n option.
The underlying method is described in detail in the JChem Database Concepts section.
Restrictions: like in the case of tautomer search option.
Explicit Hydrogens
From JChem version 5.1.3 explicit plain or isotope hydrogens in tautomerizable groups can relocate. The explicit Hydrogen constraint is enforced at the same time in the migrated location for substructure search, and full structure search, full fragment search in database (yet).
Handling of polymers and mixtures
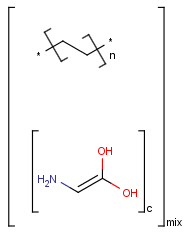
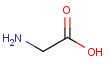
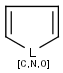
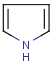
Polymers are not processed by TautomerizationPlugin, therefore their tautomers are not retrieved. E.g.:
|
Full structure search with Tautomer search |
||
|
Query |
Target |
Hit |
|
|
|
|
If polymers form a mixture with specific molecules that could have a tautomer, then these specific molecules in the mixture won't be tautomerized either because of the non-tautomerizable polymers. E.g.:
|
Full structure search with Tautomer search |
||
|
Query |
Target |
Hit |
|
|
|
|
In generic tautomer workflow searches (e.g., full fragment search), the tautomerizable fragment of a mixture containing polymer won't find the whole mixture, as the query has a generic tautomer while the target has not. E.g.:
|
Full fragment search with Tautomer search |
||
|
Query |
Target |
Hit |
|
|
|
|
Vague bond search
These search options allow to choose between several levels of strictness in matching bond types, especially regarding aromaticity. The higher the level is, the more tolerant the bond matching becomes. Vague bond options are only used when exactBondMatching is off. Otherwise (e.g. for DUPLICATE search type), vague bond level 0 (off) is used.
To fully exploit vague bond functionality, it is best to use search objects that do aromatization inside the search object: JChemSearch and StandardizedMolSearch.
Table 3. summarizes the vague bond levels focusing on aromaticity; the following sections and Table 8 describe them in detail.
Table 3.
|
Vague bond level |
Description |
|
Does not perform vague bond matching. |
|
|
(default from version 15.9.14) |
Handling of 5-membered rings with ambiguous aromaticity. |
|
(default in versions prior to 15.9.14) |
Handling of 5-membered rings with ambiguous aromaticity, 1-atom-long aromatic ring ligands and bridging bonds between two aromatic rings. |
|
All query ring bonds, 1-atom-long aromatic ring ligands and bridging bonds between two aromatic rings become ″or aromatic″ or ″any″. |
|
|
All query bonds (ring and chain) become ″or aromatic″ or ″any″. |
|
|
Ignore all bond types. |
Methods used in vague bond search
5-membered rings with ambiguous aromaticity
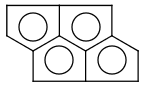
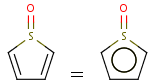
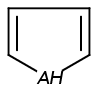
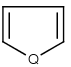
Handles some commonly occurring 5-membered query ring patterns formulated in Kekule format that have ambiguous aromaticity. This way it can return hits "visually expected by chemists", although strict bond matching would not return these. A few such ambiguous ring substructures are depicted below, with their corresponding aromatic and nonaromatic superstructures.
Table 4.
|
Ambiguous substructure |
Aromatic example |
Nonaromatic example |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
This method (used by default from JChem 3.2) ensures the expected matching of all queries where these substructures appear. On the other hand, when these rings are not not handled, query would match only the aromatic or the aliphatic targets, depending on the ambiguous query ring. (See examples.)
For efficiency reasons, above 5 such 5-membered ring patterns in the query, these ambiguous ring patterns work the same way as all ring bonds described in level 2 below.
Table 5. shows the difference between handling and not handling ambiguous rings combined with the application of different generic query atoms
Table 5.
|
Query |
Target |
|||||||
|
ambiguous aromatic rings |
||||||||
|
not handled (vague bond level = 0) |
handled (vague bond level > 0) |
|||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1-atom-long aromatic ring ligands
Used by default from JChem 5.3. Those single or 'single or double' bonds that are connected to an aromatic ring are allowed to match to an aromatic bond, except if
-
there is another ligand on the same ring atom, or
-
the bond continues in a longer (more than 1 bond long) chain
Remark: when the bond is connected to an ambiguous 5-membered ring, it can match to an aromatic bond only if the ring is evaluated to aromatic.
Table 6.
|
Query |
Target |
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Bridging bonds between two aromatic rings
Used by default from JChem 5.3. Single bonds connecting two aromatic rings are allowed to match to an aromatic bond. See also the remark about ambiguous rings at the previous method.
Table 7.
|
Query |
Target |
|||
|
|
|
|
|
|
|
|
|
|
|
|
Generalizing bond matching
Generalizes bond matching so that a bond can also match aromatic, or can totally ignore query bond types.
Vague bond search levels
Level 0 (vague bond matching off)
This corresponds to the behavior before JChem 3.2.
This method must be used if you would like to make distinction between different resonant structures, and you are passing molecules in Kekule(unaromatized) format into the search object. (MolSearch class only).
Level half (default from version 15.9.14)
Applied method:
Level 1 (default in versions prior to 15.9.14)
Applied methods:
Higher levels (vague bond levels 2-4)
The higher level vague bond options are convenience options. Their effect can be achieved also by using appropriate query bond types in the query. These options should be used during database searching carefully, because they make fingerprint screening inefficient.
They have the following effect - focusing on aromaticity - on the query bond types:
Level 2 Generalizes all ring bond types to also match aromatic.
Also applies '1-atom-long aromatic ring ligands' and
'Bridging bonds between two aromatic rings' methods
(since all ring bonds can match aromatic, all rings are considered aromatic)
Level 3 Generalizes all bond types to also match aromatic.
Level 4 Ignores all bond types.
Table 8. describes what bond type transformations are performed on the query before the search:
Table 8.
|
Original bond type in query |
Vague bond level |
||
|
2 (Ring bonds + special ligands and bridging bonds) |
3 (All bonds) |
4 (All bonds) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Abbreviations in Table 8.: S - single; D - double; T - triple; Ar - aromatic; S/D - single or double; S/A - single or aromatic; D/A - double or aromatic; A - any.
Table 9.
|
Query |
Target |
|||||||
|
|
|
|||||||
|
Vague bond level |
Vague bond level |
|||||||
|
0 (off) |
2 |
3 |
4 |
0 (off) |
2 |
3 |
4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Checking sp-hybridization state
The sp-hybridization option specifies if the sp-hybridization state of the atoms should be considered.
Calculation of the sp-hybridization state
The following states are considered:
-
sp - line configuration (e.g. C in CO2)
-
sp2 - planar configuration (e.g. C atoms in benzene)
-
sp3 - tetrahedral configuration (e.g. C in methane)
The sp hybridization state of hetero atoms is also defined by counting their lone electron pairs.
This calculated sp-hybridization state reflects the spatial configuration of the C, N and O atoms rather than the sp-hybridization of the orbitals. It doesn't cover all the mixed orbitals of Si, S and P etc. atoms.
The rules for defining the sp-hybridization state of an atom can be seen on Table 10.
Table 10. Calculation rules
|
Hybrdization state |
Conditions (OR relation) |
|
unknown |
|
|
s |
|
|
sp |
|
|
sp2 |
|
|
sp3 |
|
If checking is required, in some cases we obtain less hits than without sp-hybridization checking, because the formerly matching atoms have different sp-hybridization state.
Examples for searching with sp-hybridization checking
Table 11.
|
Query |
Target |
|||
|
|
|
|||
|
Sp-hybridization checking |
||||
|
ON |
OFF |
ON |
OFF |
|
|
|
|
|
|
|
Sp-hybridization checking may be used together with vague-bond level 4. In this case all bonds in the query match all kinds of target bonds. Using these two options molecules having atoms with the same sp-hybridization state are retrieved regardless of their bond type.
Table 12. Results with vague-bond level 4, ignoring all bond types.
|
Query |
Target |
|||||
|
|
|
|
||||
|
Sp-hybridization checking |
||||||
|
ON |
OFF |
ON |
OFF |
ON |
OFF |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Examples for searching with different implicit H matching modes
For these examples the search type is set to duplicate .
Table 13. Results with different implicit H matching modes, duplicate search.
|
Query |
Target |
Implicit H matching |
|||
|
Enabled |
Disabled |
Ignore |
Ignore and Isotope matching switched off |
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table 14. Charge matching mode ignore forces implicit H matching mode ignore in case of duplicate search.
|
Query |
Target |
|
|
|
|
Charge matching |
|
|
Ignore |
|
|
|
|
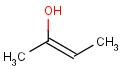
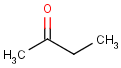


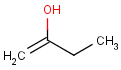
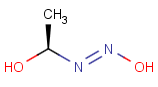
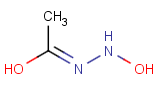
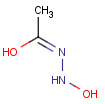
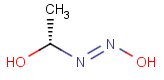
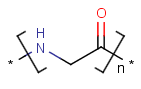
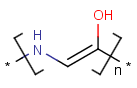
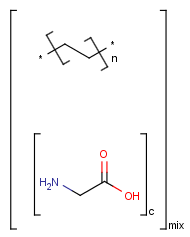
 =
= 
 =
= 

 =
= 


 =
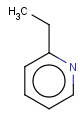
= 
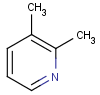 =
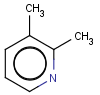
= 
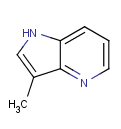 =
= 
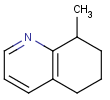 =
= 
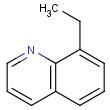 =
= 
 =
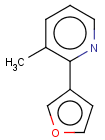
= 
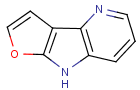 =
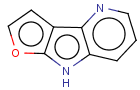
= 
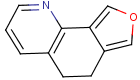 =
= 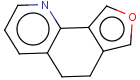
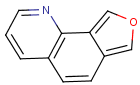 =
= 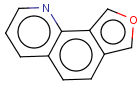
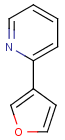
 (S)
(S)  (S/A)
(S/A)  (A)
(A)  (D)
(D)  (D/A)
(D/A)  (T)
(T)  (Ar)
(Ar)  (S/D)
(S/D)  =
= 
 =
=