Geometrical Descriptors Plugin
This manual gives you a walk-through on how to use the Geometrical Descriptors Plugin:
Introduction
The Geometrical Descriptors Plugin calculates 3D geometric descriptors for a molecule. The following window shows a calculation result.
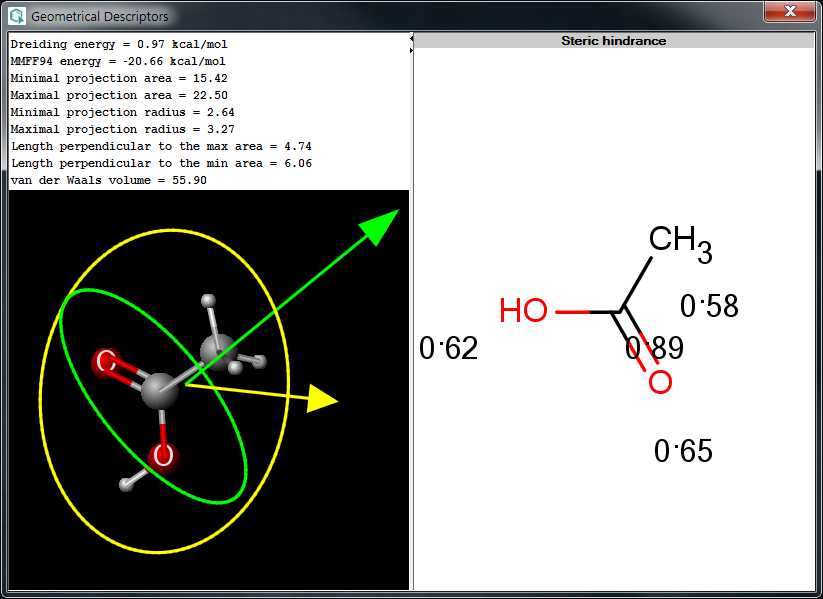
Fig.1 Geometrical Descriptors Plugin result window
Options
The following options can be set in the Geometrical Descriptors Options window:
-
Type
-
Dreiding energy: calculates the energy of the 3D structure (conformation) of the molecule using Dreiding force field.
-
MMFF94 energy: calculates the energy of the 3D structure (conformation) of the molecule using MMFF94 force field.
-
Steric hindrance: calculates steric hindrance of an atom calculated from the covalent radii values and geometrical distances.
-
Minimal projection area: calculates the minimum of projection areas of the conformer, based on the van der Waals radius (in Å2).
-
Maximal projection area: calculates the maximum of projection areas of the conformer, based on the van der Waals radius (in Å2).
-
Minimal projection radius: calculates the radius for the minimal projection area of the conformer (in Å).
-
Maximal projection radius: calculates the radius for the maximal projection area of the conformer (in Å).
-
Maximal distance perpendicular to the min projection: calculates the maximal extension of the conformer perpendicular to the minimal projection area (in Å).
-
Maximal distance perpendicular to the max projection: calculates the maximal extension of the conformer perpendicular to the maximal projection area (in Å).
-
van der Waals volume: calculates the van der Waals volume of the conformer (in Å3).
-
-
Energy unit: unit of the calculated energy (kcal/mol or kJ/mol).
-
Decimal places: setting the number of decimal places with which the result value is given.
-
Set MMFF94 optimalization: The structure is optimized before MMFF94 energy calculation.
-
Set projection optimalization The structure is optimized before projection area and projection radius calculation(s).
-
Calculate for lowest energy conformer:
-
If molecule is in 2D: the lowest energy conformer is generated if the input molecule is in 2D; 3D input molecules are considered in the given conformation.
-
Never: the input molecule is used for calculation.
-
Always: the lowest energy conformer is generated (for 3D and 2D molecules as well).
-
-
Optimization limit: sets the rigour of convergence for optimization. Stricter convergence means lower gradient limit to reach during optimization. The values can be:
-
Very loose
-
Normal
-
Strict
-
Very strict
-
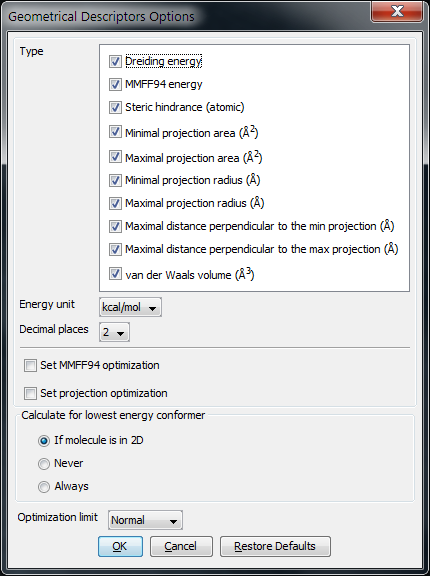
Fig. 2 Geometrical Descriptors Options window