MarvinView JFileChooser Example
This section demonstrates the possible integrations of Marvin to Swing components in a practical example.
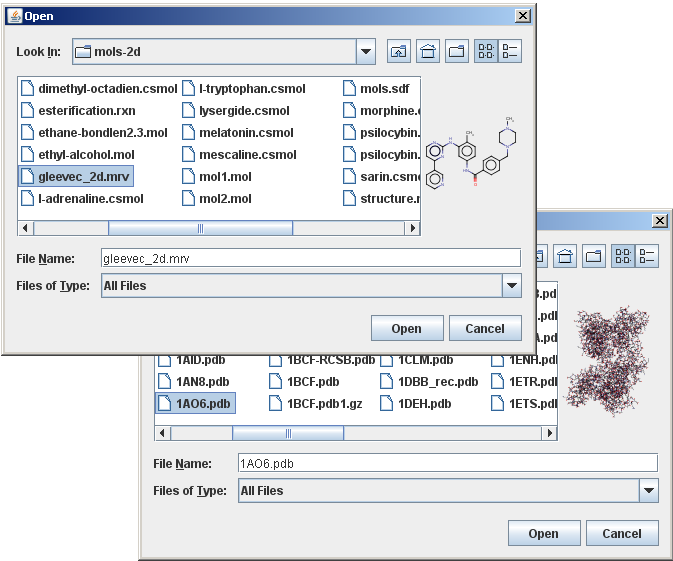
In this example we use Marvin as a custom accessory component of a file chooser to provide preview to molecule files.
In case the currently selected item in the list of the file chooser is a structure file that Marvin can import, the accessory component displays a thumbnail sketch of the molecule rendered by Marvin.
Otherwise, the accessory component is empty.
The file chooser with this custom accessory component is shown in the image below:
The following text about creating custom accessory components is quoted from the Java Tutorials:
Any object that inherits from the JComponent class can be an accessory component.
The component should have a preferred size that looks good in the file chooser.
The file chooser fires a property change event when the user selects an item in the list.
A program with an accessory component must register to receive these events to update the accessory component whenever the selection changes.
The following paragraphs describe the major steps of setting up the accessory component.
Creating and setting up the renderer
The MolPrinter class is applied to render the molecules. In case the molecule coordinates are not given in the structure file, the 2D coordinates are automatically computed.
private static class MoleculeRenderer extends JComponent { private MolPrinter mPrinter = new MolPrinter(); public void setMolecule(Molecule molecule) { // Passing the current molecule to MolPrinter. mPrinter.setMol(molecule); } public void paintComponent(Graphics g) { // It is very important to set the scale factor of MolPrinter, // otherwise the molecule will not appear. // The scale factor is computed by MolPrinter from // the current size. mPrinter.setScale(mPrinter.maxScale(getSize())); mPrinter.paint(g, new Rectangle(getSize())); }}Following file selection changes
To update the preview of the molecule in case the user selects a different file, the accessory component has to react to file selection change events with the help of a property change listener. In the example the SimpleJFileChooserAccessory object itself serves as the java.beans.PropertyChangeListener for the specific JFileChooser.SELECTED_FILE_CHANGED_PROPERTY property of the file chooser.
public SimpleJFileChooserAccessory(JFileChooser fileChooser) { molImporter = new MolImporter(); renderer = new MoleculeRenderer(); ... fileChooser.addPropertyChangeListener( JFileChooser.SELECTED_FILE_CHANGED_PROPERTY, this); } //This method handles file selection change events. public void propertyChange(PropertyChangeEvent evt) { File selection = (File)(evt.getNewValue()); if(selection != null) { try { //Passes the selected molecule file to the importer, //or the first molecule in case of a multi-molecule file. molImporter.setFileName(selection.getAbsolutePath()); //Passes the molecule from the importer to the renderer. setMolecule(); } catch (IOException exception) { renderer.setMolecule(new Molecule()); } } } //Passes the molecule from the importer to the renderer. private void setMolecule() { try { renderer.setMolecule( molImporter.nextDoc().getPrimaryMolecule()); renderer.repaint(); } catch (IOException exception) { renderer.setMolecule(new Molecule()); } }
Assigning the custom accessory component to the file chooser
The example calls the setAccessory method to establish an instance of the SimpleJFileChooserAccessory class, as the chooser's accessory component:
SimpleJFileChooserAccessory accessory = new SimpleJFileChooserAccessory(fileChooser); //Adding the accessory instance to file chooser. fileChooser.setAccessory(accessory);Sample code: SimpleJFileChooserAccessory.java
Advanced preview of multi-molecule files
Aside from a simple previewer, the accessory component can be set up to offer the possibility of browsing inside multi-molecule files without actually opening them.
The following image shows the advanced accessory component:
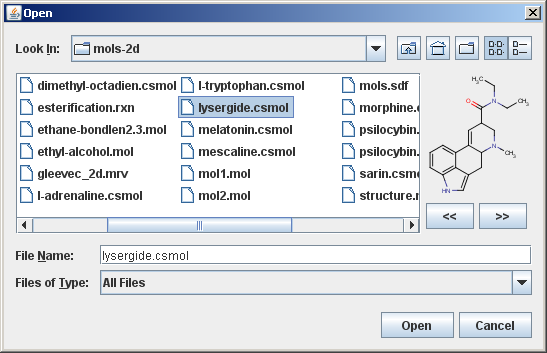
In this advanced example there are two additional buttons inside the custom accessory component that enables browsing inside multi-molecule files.
The management of the two buttons are not described in detail, only the parts closely related to Marvin, but the full source code is available.
MolImporter refers to each individual molecule in a multi-molecule file by its index (position in file, starting from 0) and provides a method to position to a molecule specified by its index.
molImporter.seekRecord(position, null);
The second argument is an optional progress monitor object which is omitted here.
The custom file chooser accessory shown in this section is already used in several ChemAxon applications and proven to be useful.