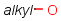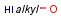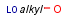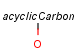Markush structure specific search options
In this document the search options concerning Markush structures are summarized and their usage is shown in different search interfaces.
Homology narrow translationThis option relates to query-side homology handling. The homology narrow translation option can switch off special handling of homology groups, hence they can match the homology group of the same type only (e.g. alkyl on alkyl or chk). The option can have the following values:
Limitations:
More information about translation options can be found in the Markush search documentation.
|
||||||||||||||||||||||||||||||||||
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE); Default value is HomologyTranslationOption.MARKED, but it is not implemented yet and works as HomologyTranslationOption.NONE. JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE); Default value is HomologyTranslationOption.MARKED, but it is not implemented yet and works as HomologyTranslationOption.NONE. Use the jc_compare operator with homologyNarrowTranslation:n/a/m Use the following command line parameter: --homologyNarrowTranslation:n/a/m |
||||||||||||||||||||||||||||||||||
|
See the availability of the option in further ChemAxon products: |
||||||||||||||||||||||||||||||||||
|
Homology broad translationHandling of target side homology groups is controlled by the homology broad translation option. If matching is switched off for a query atom, then this query atom can match homology atom only if it is the same homology atom. If matching is switched on for a query atom it matches homology group representing a larger set of structures. E.g. acyclic carbon atom can match alkyl or carbontree, an Fe atom can match transition metal or metal homology atom. Homology atoms can also match homology atoms covering a larger set of structures: carboalicyclyl can match cyclyl or xx. In this cases the query homology atom is a subset of the target homology atom. Subset rules can be seen here. Possible values for homology translation: The option can have the following vaules:
Default value is NONE. Further properties of translation NONE behavior:
Read more about homology groups.
|
||||||||||||||||||||||||||||||||||
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE); Default value is HomologyTranslationOption.MARKED, but it is not implemented yet and works as HomologyTranslationOption.NONE. JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE); Default value is HomologyTranslationOption.MARKED, but it is not implemented yet and works as HomologyTranslationOption.NONE. Use the following command line parameter:--homologyBroadTranslation:n/a/m |
||||||||||||||||||||||||||||||||||
|
See the availability of the option in further ChemAxon products: |
||||||||||||||||||||||||||||||||||
|
Markush search enable/disableSets whether targets containing Markush features should be treated as Markush libraries or not. True - if a target containing Markush features should be treated as Markush library. Default value is false. |
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE); Use the following command line parameter: --markush:n/y Default value is n. |
|
See the availability of the option in further ChemAxon products: |
|
Complete homology groupsSets if only complete structures should match on homology groups. See detailed explanation here. |
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE); Default value is true. JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE); Default value is true. Use the jc_compare operator with completeHG:y/n Use the following command line parameter:--completeHG:y/n |
|
See the availability of the option in further ChemAxon products: |
|
Markush screeningSpecifies whether screening should be used in case of Markush search. Default value is true or y. |
JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE); Default value is true. Use the following command line parameter: --markushScreening:y/n |
|
See the availability of the option in further ChemAxon products: |
|
Markush fingerprint screeningSpecifies whether fingerprint screening should be used in case of Markush search. Default value is true. |
JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE); |
|
See the availability of the option in further ChemAxon products: |
|
Hit-index typeSets the representation to use for hit indexes in case of Markush search. |
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE); Default value is SearchConstants.MARKUSH_HIT_ORIGINAL. Use the following command line parameter: --hitIndexType:m/i
|
|
See the availability of the option in further ChemAxon products: |
SearchConstants