Ward clustering
This manual gives you a description of Ward's clustering method:
Introduction
The Ward application uses Ward's minimum variance method for clustering molecules based on molecular fingerprints or other descriptors. Murtagh's reciprocal nearest neighbor (RNN) algorithm is applied as a heuristic to achieve fast calculation times.
Usage
Ward's clustering algorithm can be called as command line tool or by calling the Java class directly.
Usage as a command line tool
Use the
ward [<options>]
command to call the algorithm. You can prepare and use the ward script or batch file as described in Preparing the Usage of JChem Batch Files and Shell Scripts.
Usage as a Java class
The other way is to call the Ward class directly:
-
Under Win32 / Java 2 (assuming that JChem is installed in c:\jchem):
java -cp "c:\jchem\lib\jchem.jar;%CLASSPATH%" chemaxon.clustering.Ward [<options>]
-
Unix / Java 2 (assuming that JChem is installed in /usr/local/jchem):
java -cp "/usr/local/jchem/lib/jchem.jar:$CLASSPATH" \ chemaxon.clustering.Ward [<options>]
Because the utility has many parameters, it may be reasonable to create a shell script or a batch file for calling the software.
Options
General options:
-h --help this help message
-d --driver <JDBC driver> JDBC driver
-u --dburl <url> URL of database
-l --login <login> login name
-p --password <password> password
-P --proptable <tablename> name of property table
-s --saveconf save settings into ~/.jchem
Input options (default: standard input):
-i --input <filepath> input file path (text file input)
-q --query <sql> SQL query for reading input
(database input)
Output options (default: standard output):
-o --output <filepath> output file path (text file output)
-a --statement <sql> SQL statement for inserting results
(database output)
-x --central calculate and sign central objects
-y --singlet singletons get negative cluster ids
-z --statistics print statistics
-Z --only-statistics print only statistics
-K --Kelley <filepath> print Kelley statistics into text file
-v --verbose verbose output
Data properties
-m --dimensions <dim> number of floating-point descriptors
-f --fingerprint-size <bits> binary fingerprint size in bits
fpsize should be a multiple of 32
-w --weights <w1> <w2> ... the weights of the floating-point descriptors
-g --generate-id generate id for each compound
Clustering parameters
-c --cluster-count <count> number of clusters to be generated
-C --only-clustering clusters are generated using input RNN list
If --cluster-count is not set, then RNN list is generated on output.
Without a valid license key, the software is in demo mode and maximum 1000 structures can be retrieved from the database.
Input
The software may import data from either a text file (--input) or a database (--query). The input data must contain the following columns:
|
Columns |
Type |
Content |
|
Id |
Integer numbers |
Id of compounds |
|
fp1, fp2, fp3 ... |
Integer numbers |
Fingerprints in integer number blocks |
|
d1, d2, d3, ... |
Floating point numbers |
Other descriptors |
Comments:
-
Pharmacophore fingerprints can be generated using the GenerateMD tool. These fingerprints are not binary, so they have to be specified as other descriptors.
-
At least one binary fingerprint column or descriptor column is required.
-
Use the --generate-id option if the id column is missing from the input data.
-
Text input files can be created using the GenerateMD application. For example:
generatemd c -k CF -c cfp.xml -D < structures.smi > fingerprints.txtAn example for the XML configuration file can be found in the examples/config directory (examples\config for Windows users).
-
In the case of text input, the delimiter between two numbers should be space or tab (comma is not allowed).
-
The cd_id and cd_fpi columns in JChem's structure tables are appropriate as input.
-
In the case of database input, an SQL select statement is needed to retrieve the columns. For example:
ward -q "SELECT cd_id, cd_fp1, cd_fp2, cd_fp3, cd_fp4, cd_fp5, cd_fp6 FROM structures" ...(For the sake of readability only 6 fp. columns is applied in the above example, but usually this number is much higher.) You may also modify here the order of the results, as described in our FAQ.
-
It is important to place the query statement between quotes because it contains spaces.
Output
The software can write the results of clustering into either a text file (--output) or a database table (--statement). The exported data contains the following columns:
|
Columns |
Type |
Content |
|
Id |
Integer numbers |
Identifier of compounds |
|
Clid |
Integer numbers |
Cluster identifier |
|
Centr |
Integer numbers |
Displays whether the object is central |
The last column is written only if the --central option is specified. A central object has the smallest sum of dissimilarities to the other objects in the cluster. Central object calculation slows down the application significantly.
Comments for text output:
-
The Id and Clid columns are the same as in the case of database output.
-
A "@" symbol is used to designate the central objects of the clusters
Comments for database output:
-
A precondition of database output is the existence of a database table that contains the above columns. Create the database table before starting the calculation.
Examples for table creation:-
If the result will not contain central objects
CREATE TABLE clusters (cd_id INTEGER NOT NULL PRIMARY KEY,cluster_id INTEGER) -
If the result will contain central objects
CREATE TABLE clusters (cd_id INTEGER NOT NULL PRIMARY KEY, cluster_id INTEGER,central SMALLINT)
-
-
Before clustering, make sure that the table is empty. The SQL DELETE statement may be applied for deleting the rows in a database table. Example for deleting all rows:
DELETE FROM clusters; -
In the case of database output, an SQL statement is needed to be specified for Ward (-a option), which inserts the rows containing the results. For example:
ward -a "INSERT INTO clusters(cd_id, cluster_id, central) VALUES(?,?,?)" ...The ? symbols will be substituted with the corresponding values.
-
If the table is filled with the results, the clusters may be retrieved using SQL SELECT statements. For example:
SELECT * FROM clusters WHERE cluster_id = 1 -
It is important to place the import statement between quotes because it contains spaces.
-
The central column is 1 if the object is central, 0 otherwise
Parameters
--fingerprint-size: the number of binary fingerprint columns multiplied by 32 (because the bit-length of integer numbers is 32 in Java).
--dimensions: specifies the number of other columns. If only binary fingerprints are used in the clustering process, then this parameter doesn't have to be set.
--weights: when other columns are used, a weighted Euclidean distance calculation may be applied. If there are also binary fingerprint columns, weights are relative to the Tanimoto coefficient calculated from the binary fingerprints (the Tanimoto coefficient has a weight of 1.0).
--cluster-count: the desired number of clusters.
By default, the heap size in some Java runtime environments is limited to 64MB, so you may run out of memory easily. See the FAQ on increasing the heap size.
Saving settings
It would be inconvenient to enter all of the parameters of the ward script at each run. To overcome this problem, it is possible to save some of the settings that are not changed frequently in the .jchem file stored in the user's home directory. Use the --saveconf option to store the following settings:
-
JDBC driver's class name (--driver)
-
JDBC URL of database (--dburl)
-
Login name (--login)
-
Password (--password)
-
Binary fingerprint size (--fingerprint-size)
The settings needed for the database connection are also modified and saved by JChemManager. If you successfully entered into the database using JChemManager, then you don't need to set connection for Ward manually.
Automatic cluster level selection
Hierarchic clustering techniques, like the Ward method, can cluster the set at any chosen hierarchy level. However, in most cases, there is no obvious way to select the optimal number of clusters. Using the --Kelley <filepath> option, an optimized hierarchy level can be calculated using the Kelley method and the resulting statistics is written into the specified file.
The Kelley measure balances the normalized "spread" of the clusters at a particular level with the number of clusters at that level. For a given cluster level l, it is defined as:
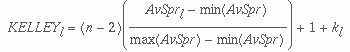
where n is the number of elements in all clusters, kl is the number of clusters, AvSprl is the average spread of the cluster at level l and min(AvSpr) andmax(AvSpr) are the minimum and maximum of this value across all of the cluster levels.
The spread of a cluster m is given by:
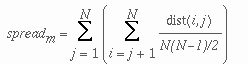
where N is the number of the members in the cluster, i and j are members of cluster m and dist(i,j) is the Euclidean distance between the two members i and j.
Running the RNN search and Ward clustering separately
Setting the --cluster-count option correctly, is important in fine tuning the clustering process. Since reciprocal nearest neighbor searching is much more time consuming than the clustering stage, it is reasonable to separate the two processes. In that case clustering can be run several times with different --cluster-count settings.
If --cluster-count is not specified, Ward collects and stores the list of RNN pairs and their distances in a text file. If this file is fed into Ward, the RNN searching is omitted. When creating the RNN list without clustering, the --common, --statistics and the --only-statistics options are not available.
If the --only-clustering option is specified for Ward, then
-
it expects an RNN list in the input text file
-
central object calculation (--central) is not available
-
the following parameters have to be specified only for the RNN calculation:
--query
--weights
--generate-id
--dimensions
--fingerprint-size
Clustering statistics
Optionally, Ward can print clustering statistics into the standard output or the given output file. The parameters that enable statistics printing are --statistics or --only-statistics. (The latter one doesn't allow to print information on individual compounds.) The following data will be printed:
-
Number of objects
-
Number of clusters
-
List of clusters (cluster id, size, central object)
-
Statistics on pairwise dissimilarity values:
-
average
-
minimum
-
maximum
-
The calculation is significantly slower if statistics is enabled, since all pairwise dissimilarity values have to be calculated. (Heuristics cannot be applied.)
Database connections
For more information on setting the following connection parameters, please visit the Administration Guide of JChem:
-
JDBC driver's class name (--driver)
-
JDBC URL of database (--dburl)
-
Login name (--login)
-
Password (--password)
Clustering examples
In the examples it is supposed that all connection parameters are set and stored by JChemManager (or a previous saving by Ward)
-
A batch file (Windows) for reading from a database and writing to the standard output:
set QUERY="SELECT cd_id, cd_fp1, cd_fp2, cd_fp3, cd_fp4, cd_fp5, cd_fp6, cd_fp7, cd_fp8, cd_fp9, cd_fp10, cd_fp11, cd_fp12, cd_fp13, cd_fp14, cd_fp15, cd_fp16 FROM structures WHERE cd_id < 10000"ward -q %QUERY% -c 100 -f 512 -
A UNIX shell script for reading from a database and writing to another table:
QUERY="SELECT cd_id, cd_fp1, cd_fp2, cd_fp3, cd_fp4, cd_fp5, cd_fp6, cd_fp7, cd_fp8, cd_fp9, cd_fp10, cd_fp11, cd_fp12, cd_fp13, cd_fp14, cd_fp15, cd_fp16 FROM structures WHERE cd_id < 10000"INSERT="INSERT INTO clusters(cd_id, cluster_id) VALUES(?,?)"ward -q "$QUERY" -a "$INSERT" -c 100 -f 512Make sure that the clusters table exists and is empty before running the script.
-
Clustering using the output of GenerateMD (in Unix):
generatemd c -k CF -c cfp.xml -D < input.smi | ward -f 512 -c 100 -g -
Clustering using pharmacophore fingerprints (in Unix):
generatemd c -k PF -c pharma-frag.xml -D < input.smi | ward -f 0 -m 210 -c 100 -g -
Testing different -c parameters. Using the output of an RNN list generation. Singletons get negative cluster ids.
generatemd c -k CF -c cfp.xml -D < input.smi > fingerprints.txt ward -f 512 -g < fingerprints.txt >neighborlists.txt ward -C -c 10 -y < neighborlists.txt >clusters.10.txt ward -C -c 50 -y < neighborlists.txt > clusters.50.txt ward -C -c 100 -y < neighborlists.txt > clusters.100.txt -
Using the Kelley method for the optimization of the number of clusters:
generatemd c input.smi -k CF -c cfp.xml -D -o fingerprints.txt ward -f 512 -g -K kelley.txt <fingerprints.txt> neighborlists.txtAn example for the generated text file (kelley.txt):
Kelley Indexes for All Cluster Levels
level index
1 500.000
2 261.018
...
18 32.038
...
498 499.000
499 500.000
Optimal number of clusters: 18Clustering using the suggested number of clusters and the generated RNN list. Singletons get negative cluster ids.
ward -C -c 18 -y < neighborlists.txt > clusters.18.txt -
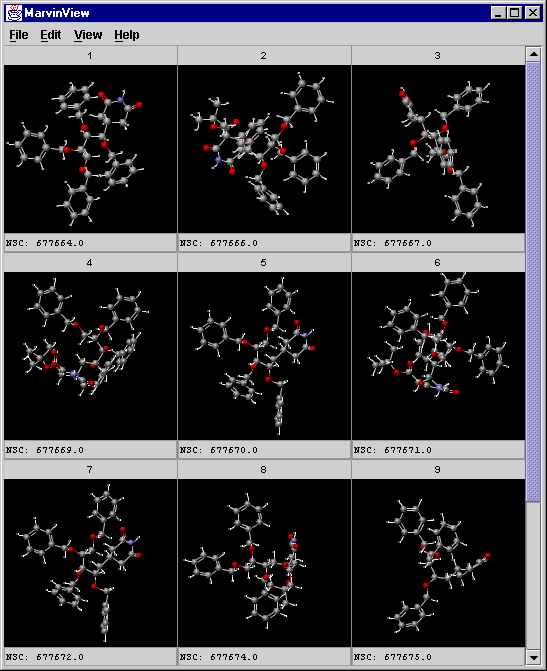
Displaying the structures of the first cluster using the CreateView and MarvinView applications:
-
Clustering:
generatemd c input.sdf -k CF -c cfp.xml -D -o fingerprints.txt ward -g -c 10 -f 512 < fingerprints.txt > clusters.txt -
Creating an SDfile containing the structures from the first cluster (clid = 1):
crview -i id -c "clid=1" -s input.sdf -t clusters.txt > ward_result1.sdf -
Displaying the structures and the NSC field (it comes from the original SDfile):
mview -c 3 -r 3 -f NSC ward_result1.sdf
A screenshot of MarvinView showing the cluster:
-
-
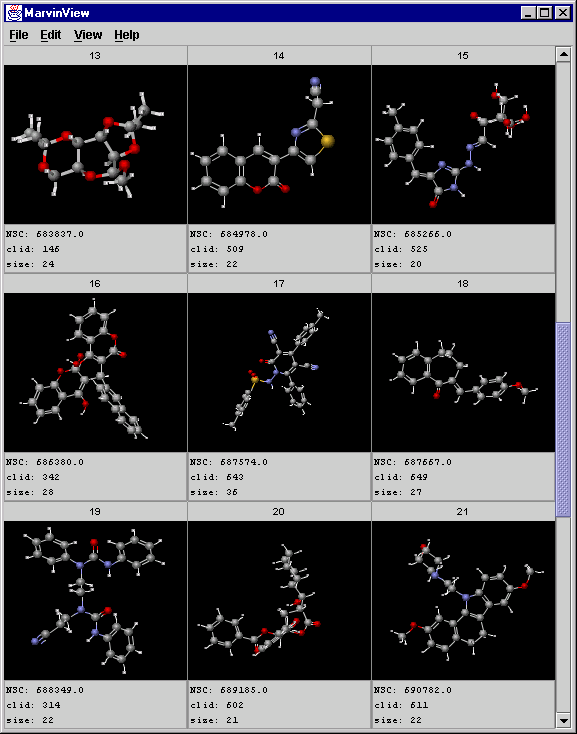
Displaying the central objects of clusters that contain at least 20 compounds (size>=20) using the CreateView and MarvinView applications:
-
Clustering:
generatemd c input.sdf -k CF -c cfp.xml -D -o fingerprints.txt ward -g -c 10 -f 512 -x -z < fingerprints.txt > clusters.txt -
Creating an SDfile containing central objects of the clusters satisfying the condition:
crview -i "centr:2" -c "size>=20" -d "clid:size" -s input.sdf -t clusters.txt > ward_result1.sdf -
Displaying the structures, the NSC field (comes from the original SDfile), and the cluster size (only for the central compounds):
mview -c 3 -r 3 -f "NSC:clid:size" ward_result2.sdf
A screenshot of MarvinView showing the central objects:
-
References
-
Ward, J. H. Hierarchical Grouping to Optimize an Objective Function J. Am. Statist. Assoc. 1963, 58, 236-244
-
Murtagh, F. A Review of Fast Techniques for Nearest Neighbour Searching. In Havranek et al. (eds.), COMPSTAT 84, Physica-Verlag, Vienna, 143-147, 1984
-
Kelley LA, Gardner SP, Sutcliffe MJ. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally-related subfamilies. Protein Eng. 1996, 9, 1063-1065