Desktop Application
The Metabolizer application enumerates the metabolites of the given substrates, and predicts their most likely major metabolites. The knowledge base for the enumeration and prediction is provided by biotransformation libraries. A human phase I xenobiotic biotransformation library is built-in the application, but you can load your own library as well.
The workflow is basically simple:
-
specify a substrate
-
start enumeration
This document describes how to use the graphical user interface.
Specifying a substrate
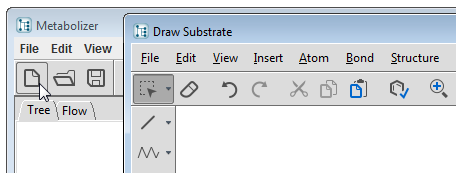
Select the File/New... menu item or click on the New... tool button  to open MarvinSketch structure editor. You can load your substrate file, draw a molecule, or paste one into MarvinSketch. When you press the Accept button of the dialog, the substrate is placed on the Tree page as the root of the metabolic tree.
to open MarvinSketch structure editor. You can load your substrate file, draw a molecule, or paste one into MarvinSketch. When you press the Accept button of the dialog, the substrate is placed on the Tree page as the root of the metabolic tree.
Enumeration
Automatic enumeration
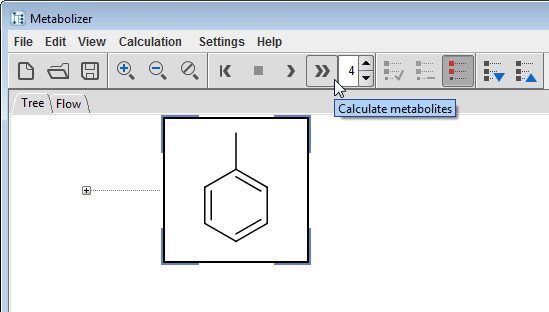
Set the Generation Limit, select a compound, and start the enumeration by clicking on the Calculation/Calculate Exhaustive menu item or its tool button  to start an automatic enumeration. The process can be stopped by pressing the Stop tool button
to start an automatic enumeration. The process can be stopped by pressing the Stop tool button  . Since this mode exhaustively produces all metabolites and their metabolites, combinatorial explosion may be a risk in case of a high generation limit. The Calculation/Calculate action
. Since this mode exhaustively produces all metabolites and their metabolites, combinatorial explosion may be a risk in case of a high generation limit. The Calculation/Calculate action  helps to decrease this risk by enumerating the major routes only. Both automatic enumeration methods can provide the same set of the most likely metabolites, the latter is just quicker.
helps to decrease this risk by enumerating the major routes only. Both automatic enumeration methods can provide the same set of the most likely metabolites, the latter is just quicker.
Manual enumeration
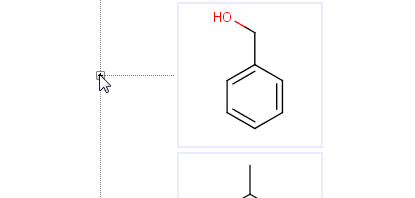
It is possible to drive metabolite enumeration manually by opening the metabolites of a compound by clicking on the plus sign before its image. The plus sign opens the corresponding branch of the metabolic tree displaying the underlying metabolites. If the metabolites are not available yet, enumeration populates the tree with the direct metabolites of the actual compound.
Reset enumeration
Existing metabolites are not re-enumerated by collapsing/expanding the nodes of the metabolic tree, but if you would like to do that, you can reset the calculations from the Calculation/Reset Metabolism menu or the corresponding tool button  .
.
Biotransformation library
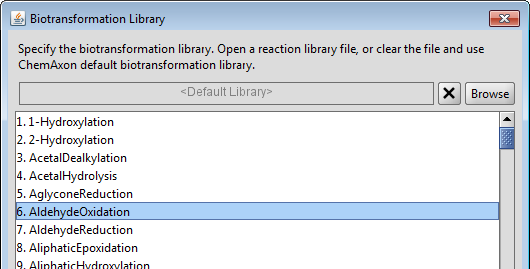
Though, Metabolizer is shipped with a buit-in biotransformation library by default, it is possible to load your own custom library even for predicting the metabolites of other species (i.e. bacterial, plant, mammal, rat). The Library Options dialog can be reached from the Settings menu. ChemAxon's reaction editor can be used to modify an existing biotransformation library or to create a new one. It allows to examine the schemes of the generic biotransformations, reactivity and selectivity rules, literature references and examples. See the Reactor documentation for more details. The Rank property is special for Metabolizer, it is a required value for major metabolite prediction. In general, the higher rank means faster reaction, but the final results are influenced by other factors as well.
Views
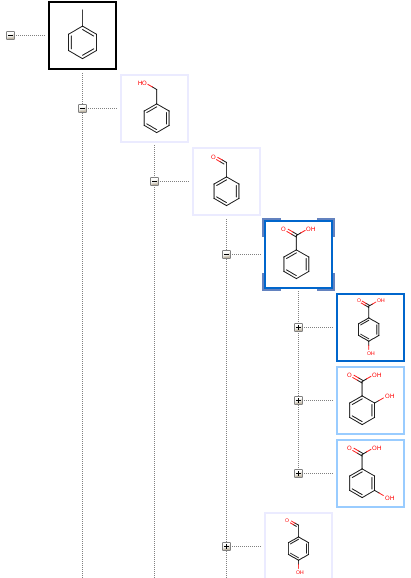
The Tree page displays metabolites as nodes of a tree having the substrate in its root on the top, and child metabolites sorted by their predicted formation rates.
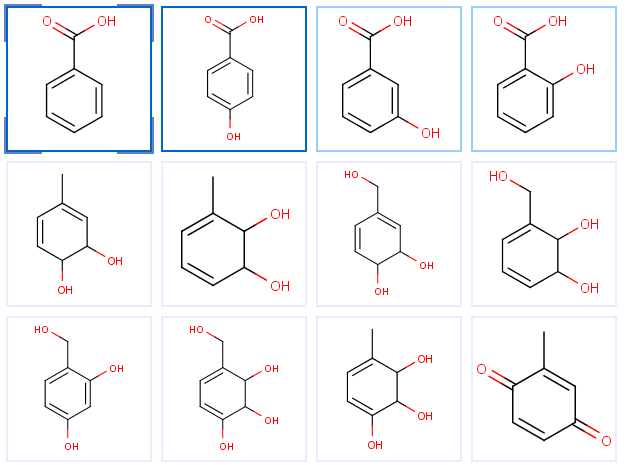
The Flow page shows metabolites sorted by their estimated global accumulation values, so the most likely metabolites are on the top of the list. Automatic enumeration can be performed on both pages, but the manual operation is possible in the Tree view only.
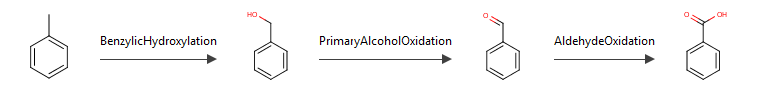
The metabolic formation route from the substrate to the selected metabolite is displayed at the bottom of both pages.
Settings
Number of generations
A limit is always set for the number of generations produced to avoid infinite execution of automatic enumeration. The optimal value depends on the substrate and the goals of the experiment, but setting this limit to 4-6 generations is the most common.
Termination condition
It is possible to specify a calculated expression to avoid the further metabolism of specific metabolites. Such conditions may limit the size or some calculable physical properties of the molecule. The Termination Condition can be specified in ChemAxon's Chemical Terms language which provides hundreds of chemical calculations. The Termination Condition dialog is available from the Settings menu.
Exclude condition
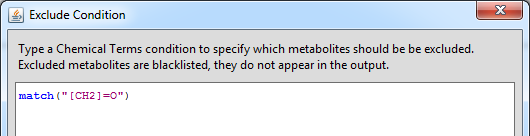
The Exclude Condition is a similar calculable expression to the Termination condition. It can serve as a black list to avoid unwanted metabolites in the output, such as small common fragments that are out of interest. The Exclude Condition dialog is available from the Settings menu.
Range options
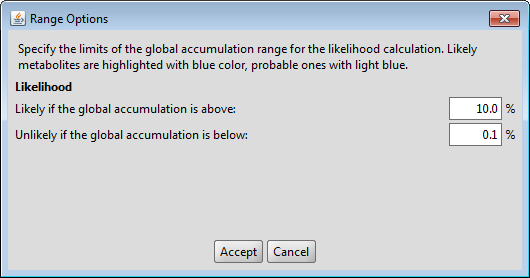
You can modify the default likelihood ranges in the Settings menu. Metabolites with high global accumulation values are considered likely and highlighted with blue color.
Glossary
Here's the collection of some often used expressions:
Substrate
The initial molecule subject to metabolic reactions.
Metabolite
A product of the metabolic reaction. It can be subject to further metabolic transformations.
Enumeration
The computational process of creating metabolites from the substrate.
Generation
Metabolites produced from a substrate by the same number of biotransformations.
Biotransformation Library
A set of generic reactions defining the transformations for the enumeration of metabolites.
Rank
Priority of a generic biotransformation (positive integer or zero).
Production
The relative amount of the original substrate transformed to a given metabolite.
Accumulation
An indicator of major metabolites calculated from production and the rates of formation and consumption reactions of a metabolites. Major metabolites accumulate faster than minor ones.