In this section search options concerning hit display are summarized andtheir usage is shown in different search interfaces.
Alignment
Specifies alignment mode of the hit structures.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setAlignmentMode( HitColoringAndAlignmentOptions.ALIGNMENT_OFF /
HitColoringAndAlignmentOptions.ALIGNMENT_ROTATE /
HitColoringAndAlignmentOptions.ALIGNMENT_PARTIAL_CLEAN );MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setAlignmentMode( HitColoringAndAlignmentOptions.ALIGNMENT_OFF /
HitColoringAndAlignmentOptions.ALIGNMENT_ROTATE /
HitColoringAndAlignmentOptions.ALIGNMENT_PARTIAL_CLEAN );
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.
alignmentMode:{off|rotate|partialClean}
jcsearch command line toolUse one of the following command line parameters to align the hits if output format is MRV.
-
--align:r rotate if query molecule has 0 dimension, it will be cleaned in 2D for alignment;
-
--align:p partial clean (template based clean) if query molecule has 0 dimension, same as rotate.
Java Server Pages
The alignment mode can be specified on the page showing the hits.
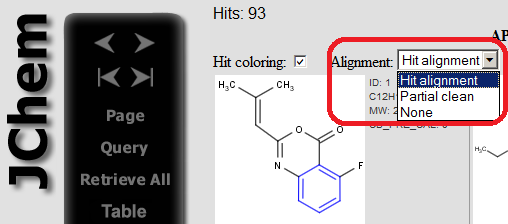
|
|
See the availability of the option in further ChemAxon products:
|
|
Coloring
Specifies whether the hit atoms and bonds should be colored or not.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setColoringEnabled( true / false );MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setColoringEnabled( true / false );
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.
coloring:{y|n}
jcsearch command line tool
Use the following command line parameter:
--hitColoring:n/y
Default value is 'n'. If the output format is MRV, colors the hits depend on search type.
Java Server Pages
Coloring can be switched off/on on the page showing the hits.
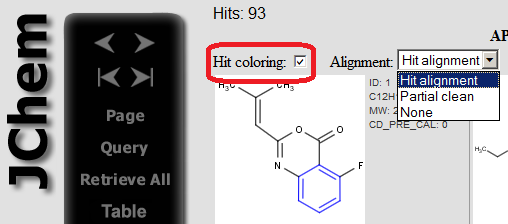
|
|
See the availability of the option in further ChemAxon products:
|
|
Hit color
Determines the color of the substructure hit.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setHitColor(Color.RED);MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setHitColor(Color.RED);
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.
hitColor:<color-value>
jcsearch command line tool
Use the following command line parameter:
--hitColor
In case of hitColoring specify color of hit atoms and bonds. Examples: "red", "green", "blue", "#00FF00"
Java Server PagesNot applicable.
|
|
See the availability of the option in further ChemAxon products:
|
Hit homology color
Specifies color of the user defined homologies in a substructure hit.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setHitHomologyColor(Color.RED);MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setHitColor(Color.RED);
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.
hitHomologyColor:<color-value>
jcsearch command line tool
Use the following command line parameter:
--hitHomologyColor
In case of hitColoring specify color of user defined homologies. Examples: "red", "green", "blue", "#00FF00".
Java Server PagesNot applicable.
|
|
See the availability of the option in further ChemAxon products:
|
|
Non-hit color
Determines the color of the target which is not part of the substructure.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setNonHitColor(Color.GRAY);MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setNonHitColor(Color.GRAY);
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.
nonHitColor:<color-value>
jcsearch command line tool
Use the following command line parameter:
--nonHitColor
In case of hitColoring specify color of non-hit atoms and bonds. Examples: "red", "green", "blue", "#00FF00"
Java Server PagesNot applicable.
|
|
See the availability of the option in further ChemAxon products:
|
|
Non-hit color 3D
Determines the color of the 3D target which is not part of the substructure.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setNonHitColor3D(Color.GRAY);MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setNonHitColor3D(Color.GRAY);
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.nonHitColor3D:<color-value>
jcsearch command line tool
Use the following command line parameter:
--nonHitColor3D
In case of hitColoring in 3D molecules specify color of non-hit atoms and bonds. Examples: "red", "green", "blue", "#00FF00".
Java Server PagesNot applicable.
|
|
See the availability of the option in further ChemAxon products:
|
Markush display mode
In case of Markush searching and hit coloring specifies the type of the resulting molecule. Ignore tetrahedral stereo
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setMarkushDisplayMode( HitColoringAndAlignmentOptions.ORIGINAL_MARKUSH /
HitColoringAndAlignmentOptions.MARKUSH_REDUCTION /
HitColoringAndAlignmentOptions.MARKUSH_REDUCTION_HGEXPANSION );MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setMarkushDisplayMode( HitColoringAndAlignmentOptions.ORIGINAL_MARKUSH /
HitColoringAndAlignmentOptions.MARKUSH_REDUCTION /
HitColoringAndAlignmentOptions.MARKUSH_REDUCTION_HGEXPANSION );
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.markushDisplayMode: <o/r/rhg>
-
o: (default) the result is shown on the given target structure;
-
r: Markush reduction to hit, the Markush structure is reduced according to the hit;
-
rhg: Markush reduction to hit and the homology groups are expanded according to the matching part of the query, which can also be a single H atom or an empty set.
jcsearch command line tool
Use the following command line parameter:
--markushDisplayMode:o/r/rhg
-
o: (default) the result is shown on the given target structure;
-
r: Markush reduction to hit, the Markush structure is reduced according to the hit;
-
rhg: Markush reduction to hit and the homology groups are expanded according to the matching part of the query, which can also be a single H atom or an empty set.
Java Server PagesNot applicable.
|
|
See the availability of the option in further ChemAxon products:
|
Remove unused definition
Sets removing unused R-group definitions in case of Markush search. By removing unused definitions readability is increased. Default value is false.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setRemoveUnusedDefinitions( true / false );MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setRemoveUnusedDefinitions( true / false );
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.removeUnusedDef:{y|n}
jcsearch command line tool
Use the following command line parameter:--removeUnusedDef:n/y
Java Server Pages
Not applicable.
|
|
See the availability of the option in further ChemAxon products:
|
Similarity
It can be applied only in case of similarity search. Describes which score is displayed within the result of a similarity search. Similarity score is displayed by default.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setSimilarityScoreDisplay( HitColoringAndAlignmentOptions.SIMILARITY /
HitColoringAndAlignmentOptions.DISSIMILARITY / HitColoringAndAlignmentOptions.SIMILARITY_OFF );MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setSimilarityScoreDisplay( HitColoringAndAlignmentOptions.SIMILARITY /
HitColoringAndAlignmentOptions.DISSIMILARITY / HitColoringAndAlignmentOptions.SIMILARITY_OFF );
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); .
JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.similarity:{s|d|o}
-
s: similarity score is displayed (default);
-
d: dissimilarity score is displayed;
-
o: neither similarity nor dissimilarity score is displayed.
jcsearch command line tool
Use the following command line parameter:--similarity:s/d/o
-
s: similarity score is displayed (default);
-
d: dissimilarity score is displayed;
-
o: neither similarity nor dissimilarity score is displayed
Java Server PagesNot applicable.
|
|
See the availability of the option in further ChemAxon products:
|
Query display
It can be applied only in case of similarity search. Describes whether query structure is displayed within the result of a similarity search. Query is not displayed by default.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setQueryDisplay( true / false );MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setQueryDisplay( true / false );
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); .
JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.queryDisplay:{y|n} (default:n)
jcsearch command line tool
Use the following command line parameter:--queryDisplay:n/y
Java Server PagesNot applicable.
|
|
See the availability of the option in further ChemAxon products:
|
|
Display labels and boxes
It can be applied only in case of similarity search. Describes whether labels and bounding boxes for the parts of the result of a similarity search - target, query, score - are displayed.
Labels and boxes are not displayed by default.
|
|
MolSearch API
HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setDisplayLabelsAndBoxes( true / false );MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
// ...HitDisplayTool hdt = new HitDisplayTool(displayOptions, searchOptions, queryMol);
Molecule result = hdt.getHit(targetMol); JChemSearch API
JChemSearch searcher = new JChemSearch();
// ...
searcher.run();
int[] idList = searcher.getResults();
List<String> dataFieldNames = new ArrayList<String>();
// ... (add required field names)
List<Object[]> dataFieldValues = new ArrayList<Object[]>(); // will store field values for the required fields
// ...HitColoringAndAlignmentOptions displayOptions = new HitColoringAndAlignmentOptions();
displayOptions.setDisplayLabelsAndBoxes( true / false );
Molecule[] results = searcher.getHitsAsMolecules(idList, displayOptions, dataFieldNames, dataFiledValues); .
JChem Oracle Cartridge
Use the jcf_hitColorAndAlign operator with hitColorAndAlignOptions.displayLabelsAndBoxes:{y|n} (default:n)
jcsearch command line tool
Use the following command line parameter:--displayLabelsAndBoxes:n/y
Java Server PagesNot applicable.
|
|
See the availability of the option in further ChemAxon products:
|

