Markush Enumeration
New to IJC 2.3 was the ability to handle databases of Markush structures. Associated with this is the ability to enumerate a Markush structure, and to restrict the enumerated structures to those that match the current query.
Markush structures are commonly used to describe combinatorial libraries or patent claims. It is assumed that the reader has basic knowledge of Markush structures.
Background
The underlying Marvin and JChem tools provide support for handling Markush structures in the following ways:
-
Marvin allows drawing and display of structures with Markush features,
-
Marvin allows enumeration of a Markush structure to generate some or all of the discrete structures described by the Markush definition. This requires a Markush Enumeration license.
-
JChem allows a table of Markush structures to be created and searches run against that table. This allows you, for instance, to perform a substructure search against a database of Markush structures to find all Markush definitions that includes the query structure as a substructure. This is very useful, for instance, when searching patent databases. Markush Search and Markush Enumeration licenses are required.
-
JChem allows Markush Enumeration to be performed within the context of a query structure, so that the structures that are enumerated are restricted to those that match the query structure. Markush Search and Markush Enumeration licenses are required. Instant JChem allows you to perform all of these operations.
1. Drawing Markush structures in Marvin Sketch
Please consult the Marvin Sketch documentation.
2. Markush Enumeration in Marvin
Marvin Sketch provides a Markush Enumeration plugin. This is a feature of Marvin, not Instant JChem, but can be used in Instant JChem whenever Marvin Sketch is used. For more details please consult the Marvin Markush Enumeration plugin documentation.
3. Creating and searching Markush tables
Creating tables is described in the Editing Entities help page.
4. Enumerating Markush structures
Instant JChem has special support for enumerating Markush structures. To do this you first need to create a JChem table containing the Markush structures.
Opening the Markush Enumeration dialog
Once you have a Markush structure table you can view the contents using the standard form or grid view. You can also run structure searches (most typically substructure searches) against this table to find only those Markush structures of interest. When you are viewing the contents of the Markush table in the grid or form view you can choose to enumerate any particular structure. Select the structure you want to enumerate and click on the 'Enumerate a Markush Structure' icon (  ) in the toolbar. The Markush Enumeration dialog will open.
) in the toolbar. The Markush Enumeration dialog will open.
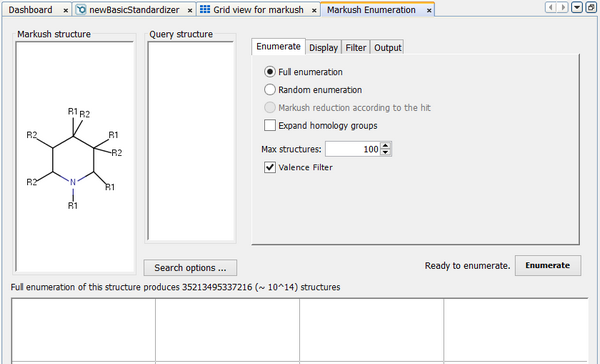
Structures
Structures are displayed in 3 regions of the window:
-
Markush structure: This is displayed in the top left. The Markush structure that you selected from the structure table is displayed, but you can edit this by double clicking on the panel.
-
Query structure: This is displayed in the panel to the right of the Markush structure. It may be empty if no structure search has been run. You can edit this by double clicking on the panel.
-
Enumerated structures: These are displayed in the main panel to the bottom of the window and will initially be empty. Once you have enumerated some structures each one can be opened by double clicking on it. This opens Marvin Sketch and allows access to the full Marvin functionality. For instance, you can continue enumeration in the case of partially enumerated structures, or you can calculate molecular properties for any particular structure.
Enumerate tab
The Markush Enumeration dialog operates in 3 different modes which can be specified on the 'Enumerate' tab:
Full enumeration
-
This performs exhaustive enumeration of the Markush structure. Markush libraries can potentially be vast in size (bigger that the number of atoms in the universe!), so the enumeration is limited to a maximum number of structures that you can specify. By default this is set to 100 structures.
Random enumeration
-
This performs random enumeration of the Markush structure. This is most useful for large Markush libraries where it is not practical to fully enumerate the library. Random enumeration allows you to sample the library in a random fashion so that you obtain a good representation of the various structures in the library. The same warning about library size that are described for full enumeration also apply to random enumeration.
Markush reduction according to the hit
-
This option is only active when you have run a substructure search on the Markush table and when you have a Markush Search license. In this mode the enumerated structures are limited to those that contain the substructure. Whilst this usually significantly reduces the number of enumerated structures, the limits on the enumerated library size still apply. You can see the part of the enumerated structure that corresponds to the query substructure using the typical hit display options.
Multiple enumerated paths matching the substructure may result in the same enumerated structure, so the results may contain duplicates.
Expand homology groups
-
This options specifies whether homology groups (Alkyl, Aryl etc.) in the Markush structure should be enumerated with examples of the group.
This gives a representative sample of structures for the homology group, but not necessarily an exhaustive set.
Max Structures
-
This limits the number of enumerated structures that are generated.
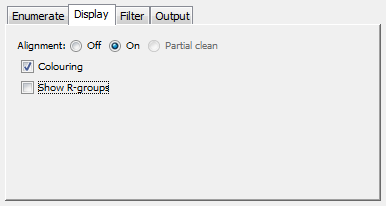
Display tab
This specified display options for the enumerated structures.
Alignment: Are enumerated structures aligned to the Markush core (full or random enumeration) or to the query structure (Markush reduction).
Colouring: Are enumerated structures coloured according to their R-groups (full or random enumeration) or to the query structure (Markush reduction).
Show R-groups: Are R-groups displayed in the Markush structure and the enumerated structures.
Display tab:
Filter tab
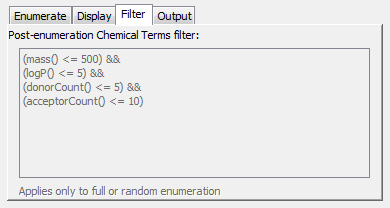
This allows a Chemical Terms filter to be specified that is applied to each enumerated structure. Those enumerated structures that fail the filter are discarded. When a filter is set you may then see fewer structures than you set as the maximum as some may be discarded. To set a Chemical Terms filter double click on the panel and the editor opens. This is the same editor that is used when specifying a Chemical Terms filter for queries , and the filter specified must return a boolean value (true or false). This filter can be useful for filtering out non-drug-like structures or similar purposes. Note that it only applies to Full or Random enumeration as Markush reduction can generate partially enumerated structures for which properties cannot be calculated. For more information about specifying Chemical Terms filters see here.
Filter tab:
Output tab
This tab lets you specify how you want your enumerated structures output. The default is to output them to the display area in the lower half of the window, in which case you can also specify the grid size. Alternatively you can output to a file. When this option is set you are prompted for the filename when you start the enumeration.
Other features
Library size - The full enumerated size of the library is displayed beneath the Markush structure. This helps you decide whether to use full or partial enumeration, and whether to adjust the limit on the maximum library size.
The actual number of enumerated structures may be less than the calculated full enumerated library size. This is because the actual enumeration includes a valence filter that excludes incorrect structures. For instance this can happen when using query bond features e.g. an ANY bond attached to a benzene ring will give a predicted library size of 3, but when the actual enumeration is performed only a single structure will be generated as the double and triple bond variants would result in valence errors.
Performing enumeration
Once the appropriate options have been set the enumeration can be started by pressing the 'Enumerate' button. Once running this button changes to 'Cancel' allowing the enumeration to be halted at it's current position. Results are displayed as the structures are generated. If enumerating to file sample enumerated structures are displayed as the enumeration proceeds.
Memory usage: Enumerated libraries can be very large. Enumeration can be slow and use lots of memory. If you are wanting to enumerate large libraries then consider:
-
Increasing the amount of memory available to Instant JChem. See the memory usage documentation for details.
-
Outputting the results to file rather than displaying them.