Scaffold Based Enumeration
The scaffold based enumeration functionality of Plexus Design is an effective tool for creating virtual compound libraries in a few easy steps.
Scaffold based (or Markush) enumeration lets you generate a combinatorial library via substituent variation resulting in virtual molecules having the same core structure (i.e., scaffold).
In the scaffold compound, those positions where substituent variation can occur are occupied by R-group atoms (see the leftmost bubble).
Fragment definitions for each R group on the scaffold should also be provided. These fragment definitions are special structures as they contain a so-called attachment point that indicates which part of the fragment will be directly connected to the core structure.
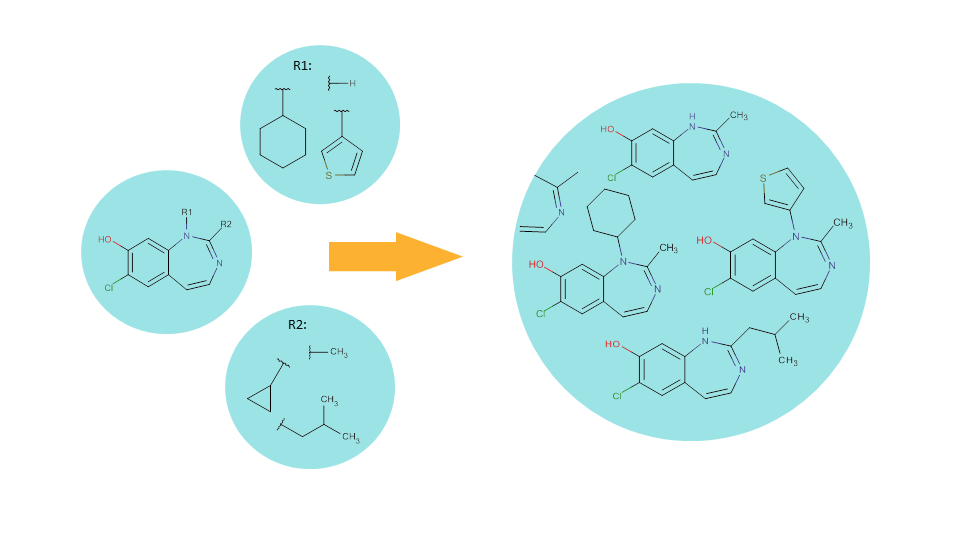
Enumerating the full library corresponds to systematically generating of all possible combinations of the R-groups on the given scaffold. As a result, we will have a virtual library that contains structures derived from the same scaffold but having different substituents.
Below, you can find a detailed description about each step required for setting up and running a scaffold based enumeration.
Preparing for a scaffold based enumeration
Scaffold based enumeration is available from the menu bar, under the Enumeration option. Once you have selected this menu item, you will be navigated to the Scaffold Enumeration page:
Creating the scaffold
The first step in the preparation for an enumeration is to define the scaffold, i.e. the common part of the later enumerated compounds. If you click in the scaffold window, the Marvin JS editor will open, where you can either draw your scaffold structure or import it from file. Your scaffold has to contain one or more R-atoms, which will be substituted by different atoms or groups during the enumeration. You can learn more about drawing structures with R-atoms in the Marvin JS User Guide.
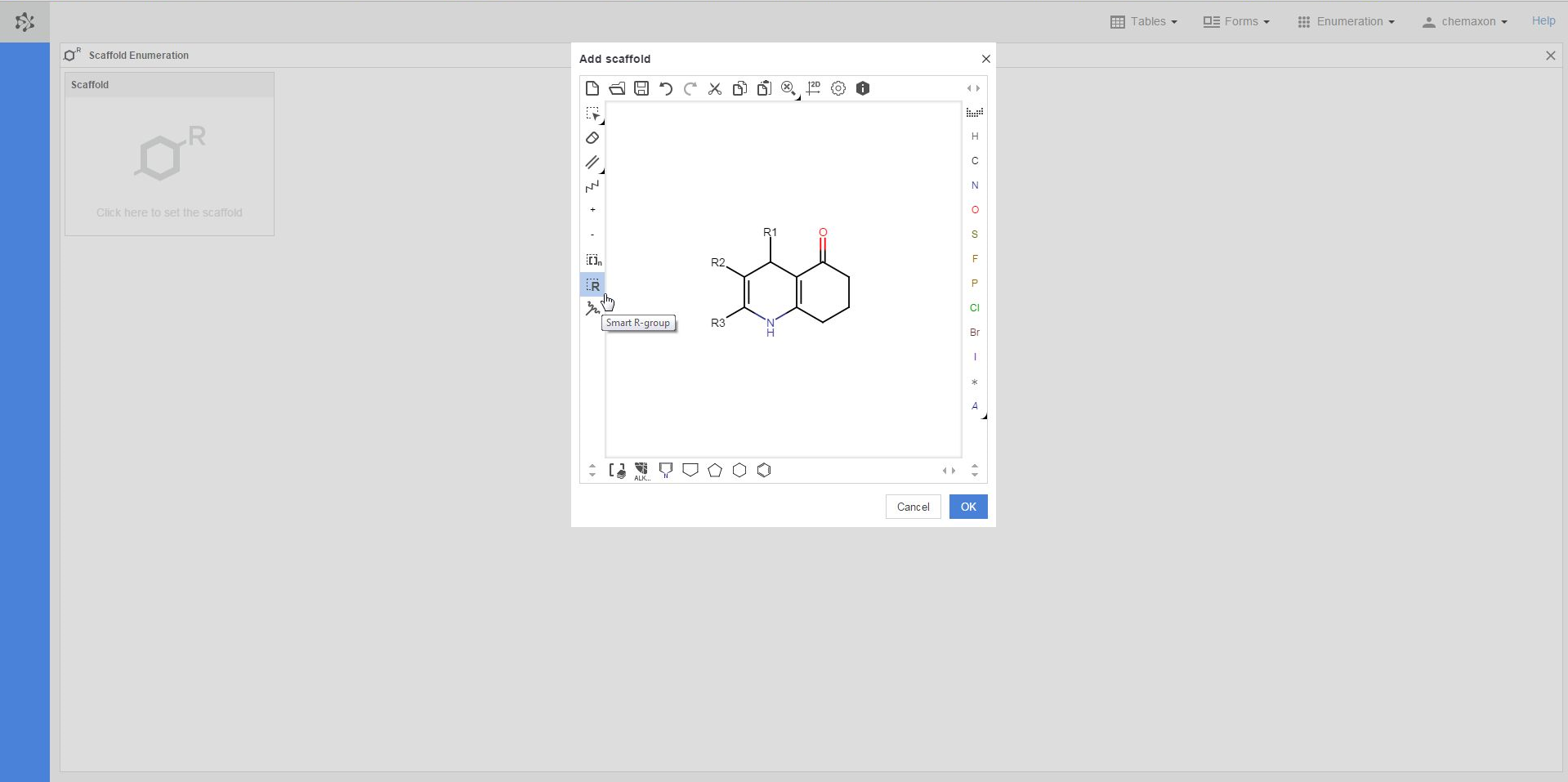
Once you have completed drawing the scaffold in Marvin JS, you can send it back to the Scaffold Enumeration page by pushing the OK button. As a result, additional boxes will appear on the page: each box belongs to a different R-atom of the scaffold molecule, and will have to contain the R-group definition for that R-atom.
A structure in an already existing table or form can also be selected as scaffold: when you right-click on a compound and select Use in scaffold enumeration from the context menu, you will be re-directed to the Scaffold Enumeration page where the chosen structure has already been set as scaffold.
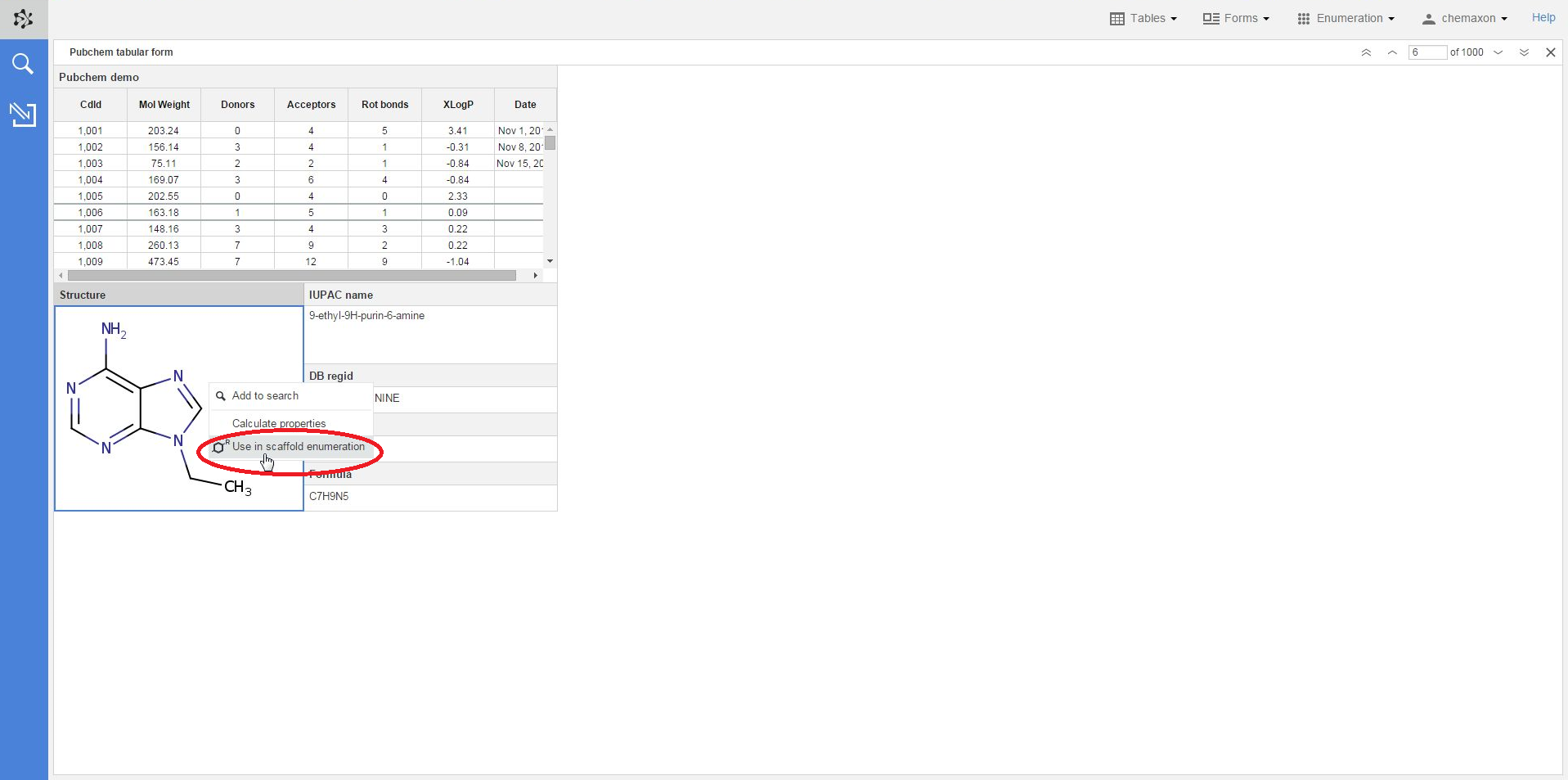
In order to use the selected molecule as a scaffold, you should add the variable parts to the structure by adding R-groups. Plexus Design then automatically recognizes the number of R-groups on the scaffold and asks you to add the corresponding fragment definitions in the next step.
Adding R-group definitions
You can create the definitions for the R-groups in three different ways:
-
Load the R-groups from collection: The R-group definitions can be imported from the Building Blocks Collection. (For creating structure lists in this collection, go to .../building-blocks, for example https://ps-demo.chemaxon.com/#/building-blocks)
-
Load the R-groups from file: The R-group definitions can be imported from the common chemical file formats which support R-group attachment points, e.g., mrv, sdf, cxsmiles.
-
Draw the R-groups: The substituents are drawn one by one in the Marvin JS editor.
These options become available once you have clicked on the "+" sign before the box of the respective R-group.
You also have to add "attachment points" to your substituents to mark the positions where the group will connect to the scaffold. Make sure that each substituent group has exactly the same number of attachment points as the number of bonds belonging to its respective R-atom in the scaffold. In the case of single atoms, defining the attachment points manually is not necessary because the application can add them to the scaffold automatically in the required number.
A more detailed description of drawing R-group definitions can be found in the Marvin JS User Guide.
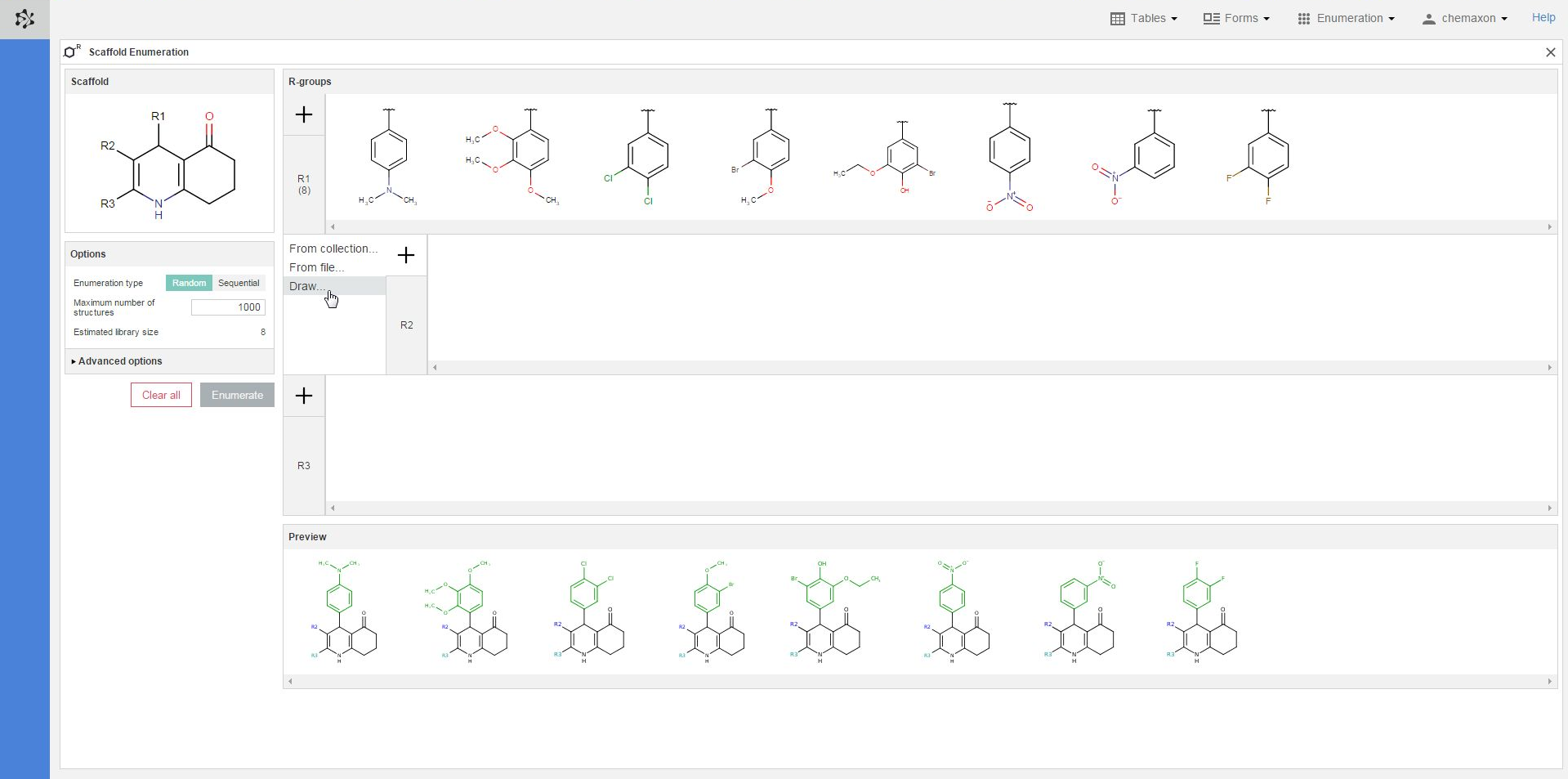
When you right-click on the structures in an R-group definitions, you will find the following items in the pop-up menu:
-
Edit: you can open the Marvin JS editor again to edit the selected structure;
-
Remove Selected: you can delete each selected substituent (one in the image below);
-
Remove Unselected: you can delete each unselected structure (8 in the image below);
-
Clear all: you can delete every substituent in the R-group of the selected structure (in the R2-group in the image below).
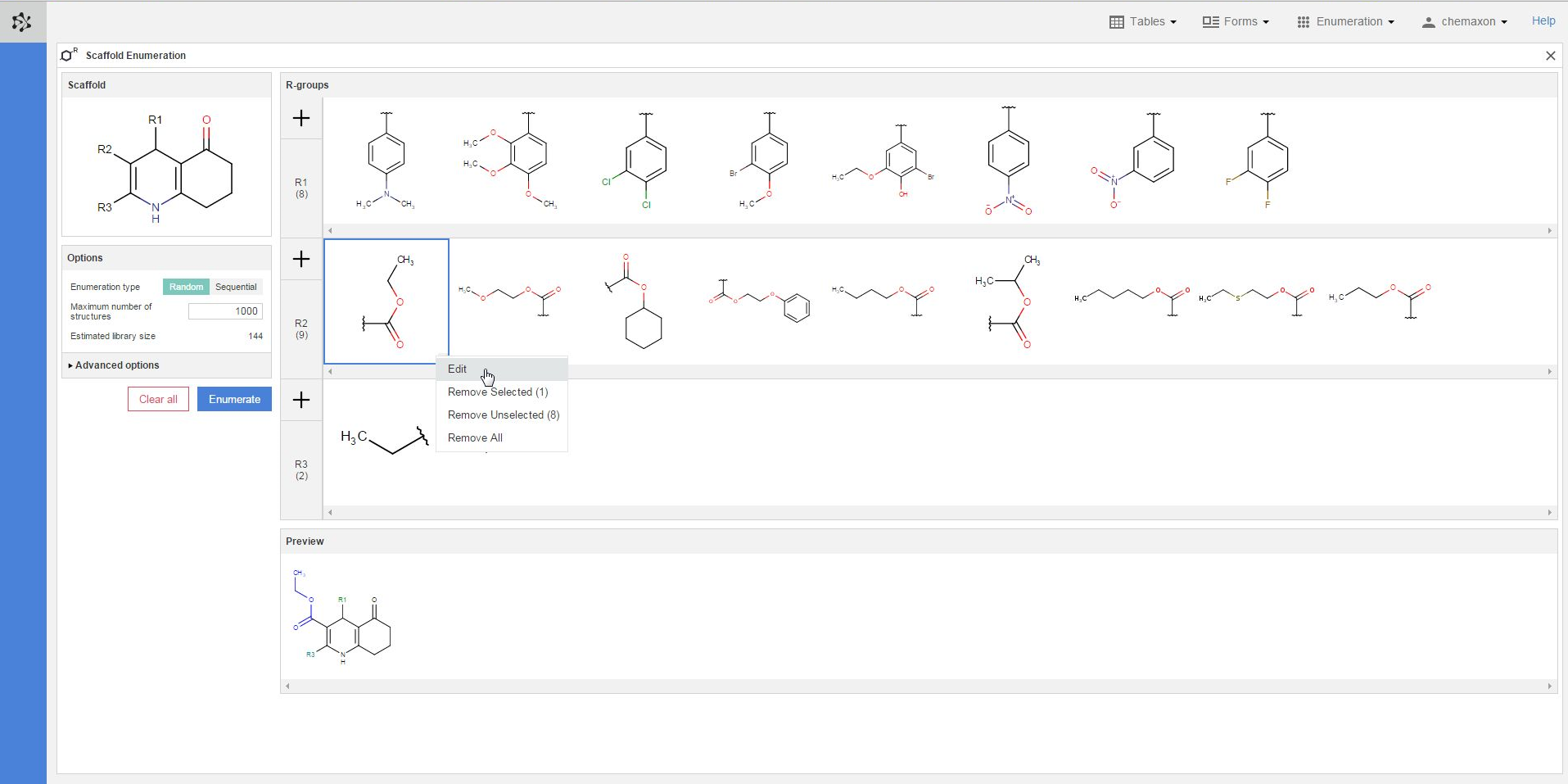
Plexus Suite analyzes the fragments when they are added to an R-group, and gives a red frame to those structures where it detects any problems. In the image below, you can see an example where the number of attachment points does not match the number of bonds belonging to its respective R-atom. The exact error message becomes visible as well if you hover over the erroneous fragment with the cursor. Similar error message appears in those cases when the bond order (the type) of the attachment point does not fit to its R-atom in the scaffold.
Enumerating Markush structures
Plexus Suite is capable of interpreting complete Markush structures (scaffold along with R-group definitions) and use them for a scaffold based enumeration. You can upload an .mrv structure file containing one or more Markush structures to Plexus Suite in order to create a new database table from the structures. Although the Structure column of this table contains only the uploaded backbone structures and not the substituents in the R-groups, once you right-click on a structure and select the 'Use in scaffold enumeration' option from the pop-up menu, you will be navigated to the Scaffold Enumeration page where the elements of the selected Markush structure have already been organized: the backbone structure has been added to the scaffold window while each substituent has been added to the appropriate R-group box.
Similarly, the scaffold and the R-groups can be created in one single step from a Markush structure file if you open the Marvin JS editor to set the scaffold and then import a file containing a single Markush structure into the editor. When you push the Add button, both the scaffold and the R-groups will be added to their respective boxes.
Setting additional options
Besides defining the common backbone of your compound library and its possible substituents, you can use further restrictions on your enumerated library.
In the Options panel of the Scaffold Enumeration page, you can decide what kind of enumeration you want to carry out:
-
Enumeration type:
-
Random enumeration: during the compound library generation, the scaffold R-atoms are replaced by randomly selecting from the available substituent combinations.
-
Sequential enumeration: the compound library is generated by systematically substituting the R-atoms with the possible groups.
-
-
Maximum number of structures: you can define the number of compounds to be generated.
The Estimated library size field, in the last row of the Options panel displays the total number of compounds which can be generated from the current set of scaffold and R-groups.
Further settings can be found when you click on the Additional options header. You can switch on or off the following four options:
-
Enumerate homology groups: Homology groups are R-groups, represented as pseudo atoms - with the names covering a set of predefined R-groups. If Off, the homology groups are kept as pseudo atoms. This might be useful for showing that these structures cannot be fully enumerated.
-
Duplicate filtering of results: Ensures that only unique molecules are saved from the results.
-
Filter valence errors: If the Markush structure is not properly (or is too generally) formulated, it is possible that it describes structures with valence errors. In this case the valence filter setting is useful to filter out the offending result structures.
-
Filter geometrically unfeasible rings: If turned On, it rejects molecules with trans double bonds in bridged rings, cumulated double bonds in rings, triple bonds in rings.
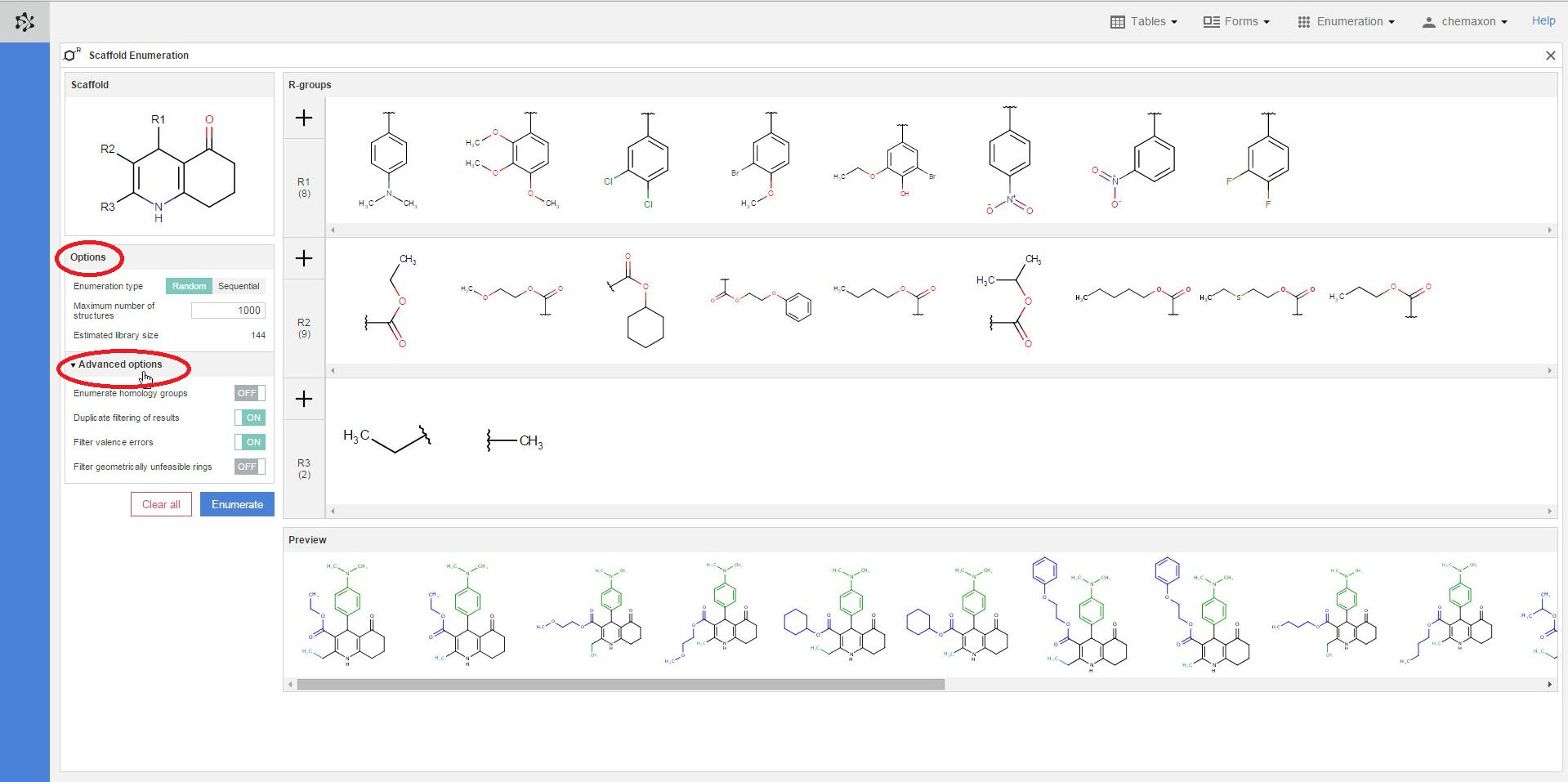
Preview of the compound library
In the preview pane, you can see a few of the molecules generated from the scaffold and the combinations of R-group definitions (substituents). The coloring of the preview molecules immediately reflects their constitution: the scaffold part is colored black, while each functional group substituting a different R-atom has its own coloring.
You can also select different substituents from each R-group definition to check some of the compounds which will be generated when the enumeration will be executed. You can select multiple items by holding down the Ctrl key; the selected substituents will get a blue frame. On the image below, two substituents were selected from the R1-group, three from the R2-group and one from the R3-group. The resulting six structures are displayed in the preview pane, where the scaffold part is black, the R1-substituents are green, the R2-fragments are blue while the R3-group is cyan:
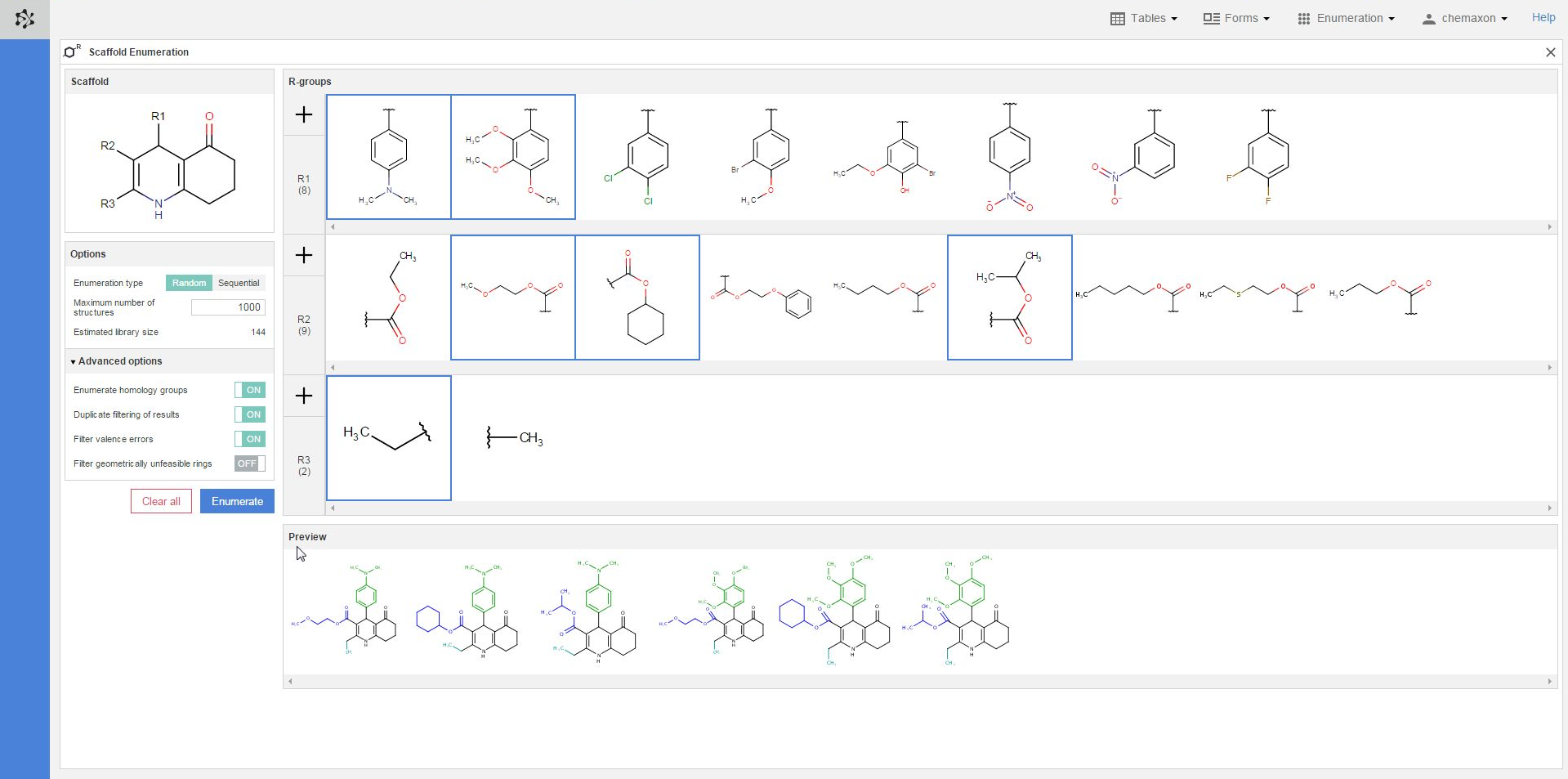
Executing the enumeration
When you have finished drawing the scaffold and the R-groups, as well as have set the options you needed, the actual enumeration can be started with the blue Enumerate button. You can also delete the whole content of the Scaffold Enumeration page by pressing the Clear all button. As a result, the scaffold and the R-group definitions will be deleted, and every option will be set to their respective default value.
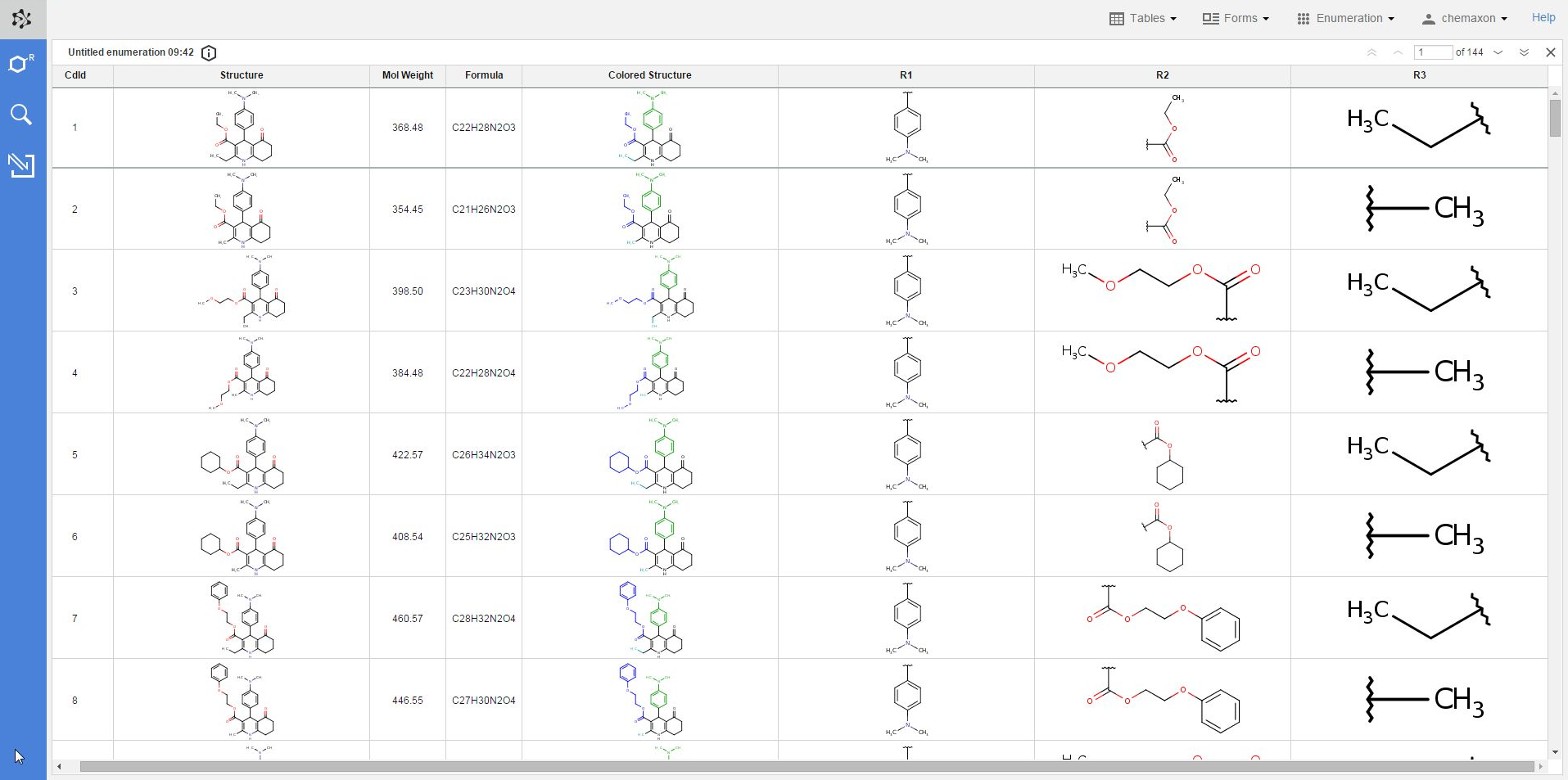
The result of each enumeration will be added as a new table to your database. Please note that just like in the case of imported files, the new table will be visible and available only from your own user account. In order to make it available for other users as well, you have to share it with one or more user roles in Instant JChem. You can read more about sharing database items with Instant JChem here.
The new table will contain the following columns: the compound identifier (CdId) of the enumerated molecules, the structure, mol weight and formula of the enumerated structure, the respective substituent group for each R-atom, and the "colored structure", which corresponds to the coloring of the enumeration preview: the scaffold has black color while each R-atom substituent appears with a different color.
Metadata in enumerated tables
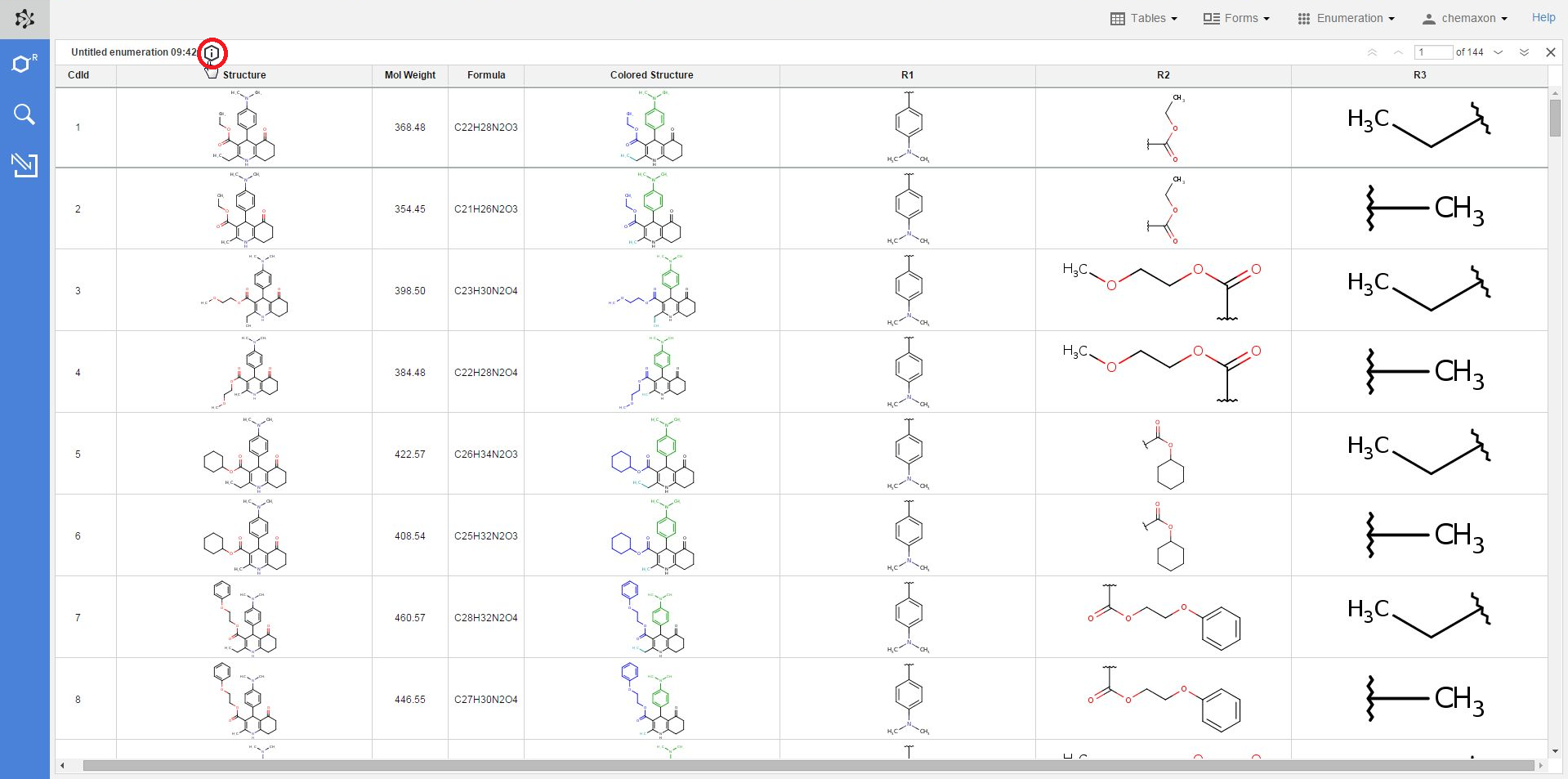
Whenever a new table has been created via enumeration, the settings used in the library design process are added to the table as metadata. With the help of this metadata, it becomes very straightforward to apply modifications to prior enumerations, and then create new libraries with these modified settings.
By default, the metadata remains hidden when you open an enumerated table, but you can open it by clicking on the information icon over to the table:
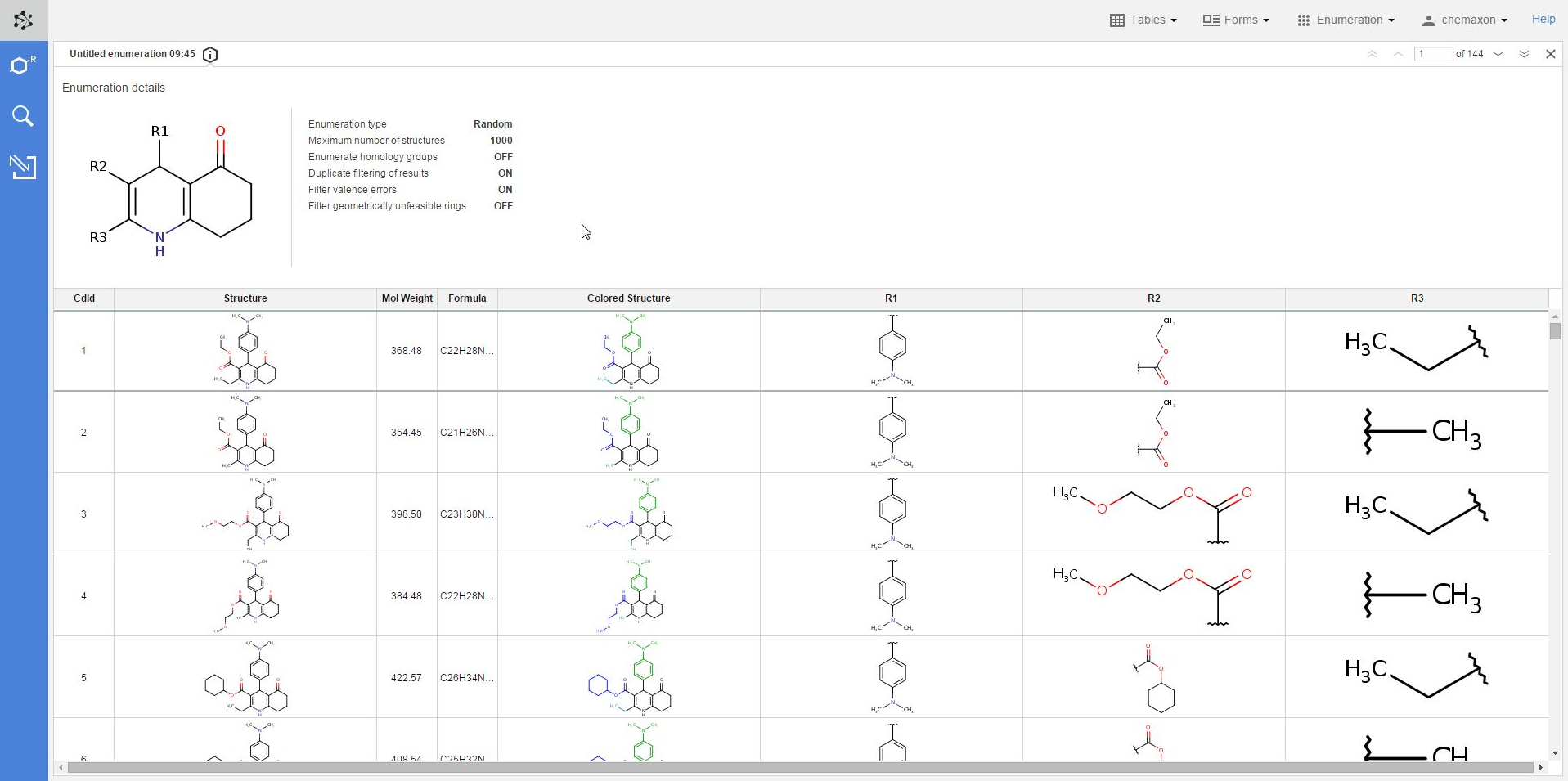
The panel sliding in contains all of the details about the enumeration which was used to create the current table, including the scaffold structure and each option setting.
A Markush icon on the top of the blue action bar is available any time you open the cretaed virtual library in Plexus Suite. Once you click on the icon, the scaffold enumeration page re-opens with the same pieces of data which were used to generate your compounds. This way, you can easily make modifications in your enumeration settings and re-enumerate your library.
Need more help? Check out our tutorial video about how to perform scaffold-based enumeration!
Visit other Plexus Suite pages to get to know the other enumeration method in Plexus Suite and to learn how the enumerated structures can be used further: