ChemCurator Installation and System Requirements
Contents
System requirements
Operating system
-
Windows 7 SP1 64 bit (or newer)
-
OS X 10.9 Mavericks (or newer)
-
Ubuntu Linux 14.04 (or any newer compatible distribution)
-
Microsoft Windows Server 2012 (or newer, for servers and cloud hosted installation)
Oracle Java 8 64 bit (Bundled into Mac and Windows installer)
|
|
Minimum |
Recommended |
|
CPU |
Dual Core 1.5+ GHz |
Quad Core 2+ GHz |
|
Free RAM |
2 GB or more |
4 GB or more |
|
Hard Disk space |
512 MB or more |
1 GB or more |
|
Screen Resolution |
1024x768 |
1920x1080 (dual screen) |
Installation
In order to download the ChemCurator installer, go to www.chemaxon.com and select ChemCurator from the Download menu. Download the proper installer to your computer.
After the download is complete, run the installer on your computer.
An Installation Wizard opens, which guides you through the installation process.
On the next screen, you can read the License Agreement. Accept this agreement in order to continue.
Then the default installation library for ChemCurator will appear.
You can modify it by clicking on Browse. After specifying your destination directory, click Next.
Components to be installed can be reviewed before starting the installation process.
Then Setup installs ChemCurator on your computer. The installation may take a few minutes.
After installation has been finished, a closing screen appears.
Click Finish to exit the Setup Wizard.
Now you are ready to start the ChemCurator application by double-clicking on the appropriate icon on your desktop. You may need to install the corresponding license, in this case please follow the steps described here.
Configuration
Depending on your working environment, some functions of ChemCurator may need additional configurations.
Automatic update
New version of ChemCurator released every week with new features, improvements and bugfixes. Automatic update function periodically checks available version updates and after user confirmation downloads and install them. Depending on user preferences automatic update can turn off, or in-house update site can be used instead of official ChemAxon update side. Automatic Update settings are available from Tools>Plugins Settings tab. The update check also can be launched manually from Help>Check for Updates.
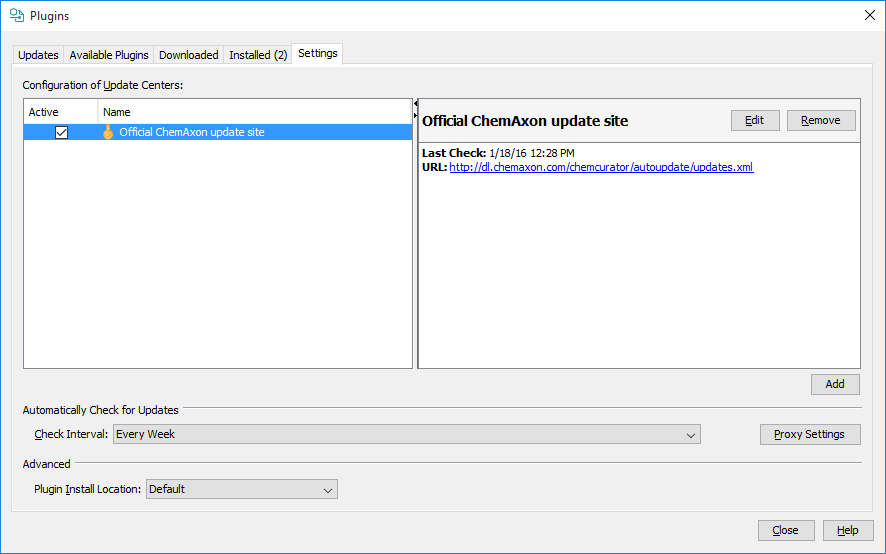
Automatic Update Configuration
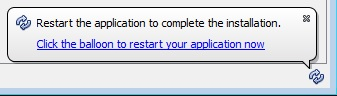
Update Notifications
Proxy settings
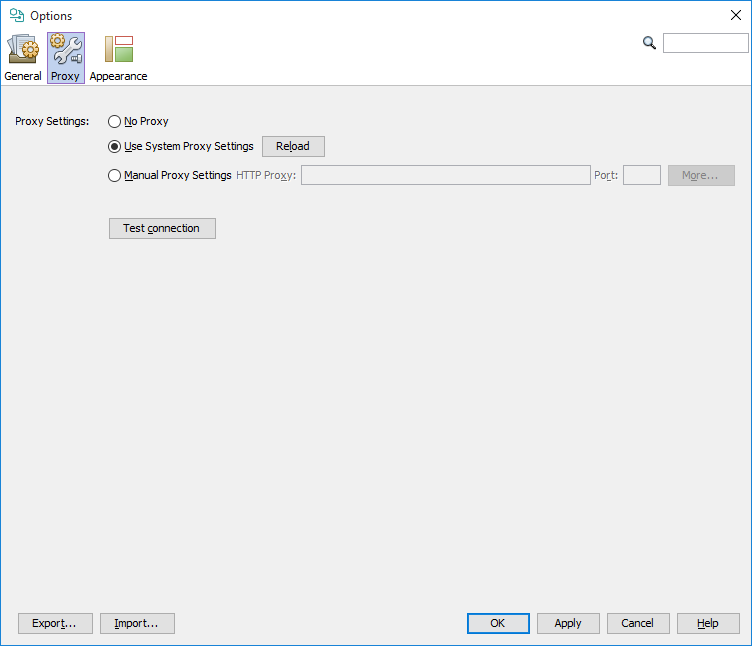
If you use the application behind a proxy server, you may need to set the proxy settings for outgoing communication. These settings are necessary for direct import from Google Patents and IFI Claims, automatic update and some other functions. You can test your settings using the Test connection button. If neither of No Proxy or Use System Proxy Settings works for you, please consult your system administrator about the proper Manual Proxy Settings. Proxy settings are available from Tools>Options... Proxy tab.
PDF annotation support
PDF annotation currently out of the box supported on Windows, on Mac and Linux you need to install Pdf2htmlEX manually.
Linux (Ubuntu)
The second and third lines era optional depending on your system configuration but usually necessary.
sudo add-apt-repository ppa:coolwanglu/pdf2htmlexsudo apt-get remove libfontforge1sudo apt-get install libfontforge-devsudo apt-get install pdf2htmlexMac
You can install Pdf2htmlEX with Homebrew . The first line is optional it is necessary only if you don't have installed Homebrew.
ruby -e "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)"brew install pdf2htmlexOSR support
If you want to recognize not only chemical names but also image based chemical structures with ChemCurator, you need an OSR (Optical Structure Recognition) application. Currently CLiDE Batch, OSRA and Imago supported.
OSR is time-consuming, which means the document annotation process can slow down with OSR.
A special CLiDE4ChemCurator version of CLiDE on Windows systems installed automatically together with ChemCurator. The bundled CLiDE can be activated without further installation with the proper license key directly from Tools>Options... General tab CLiDE sub-tab.On CLiDE subtab (you are also able to set the separately installed CLiDE location manually here). After CLiDE activation please restart ChemCurator.
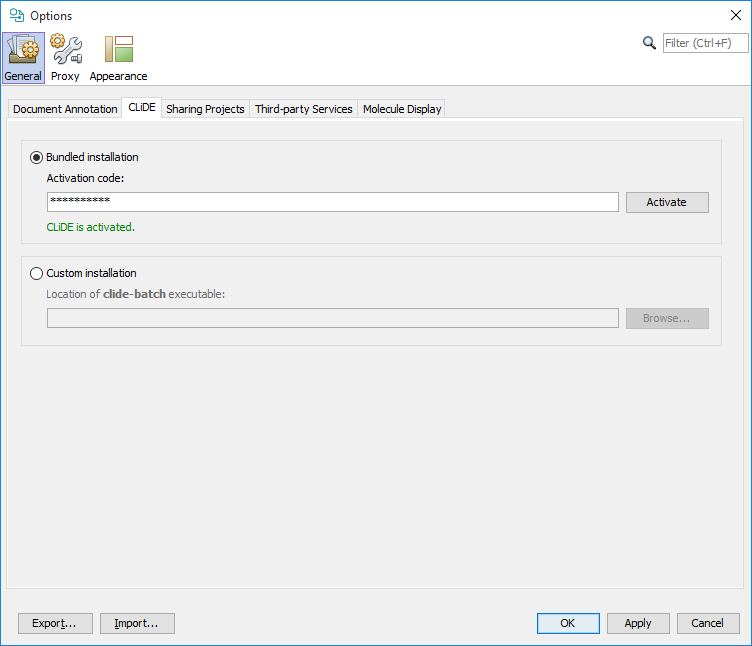
For configuration of other supported OSR applications check the corresponding section of the Document to Structure User's Guide.
Integration Server configuration
For project sharing through integration server you need to set your central Integration Server URL address and a valid username and password on Tools>Options... General tab Sharing Projects sub-tab.
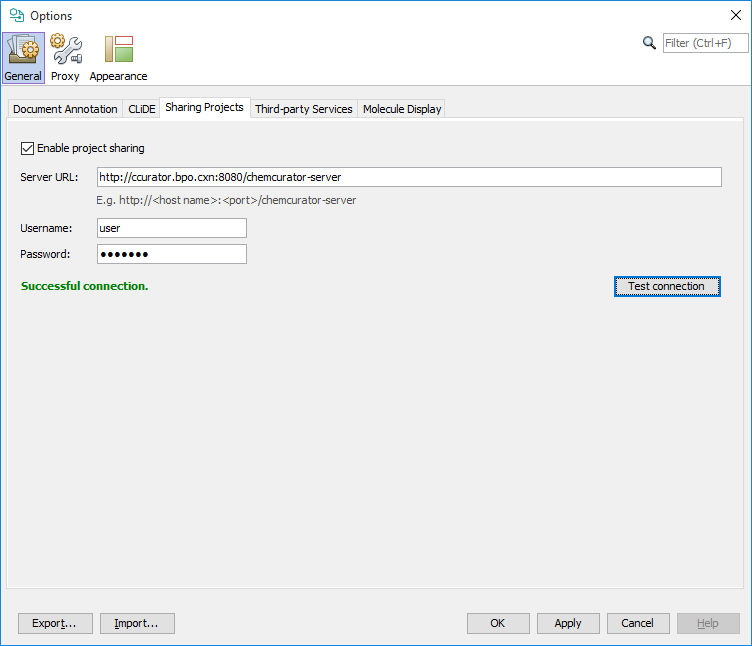
Java memory options
The application will try to allocate 2 GB of system memory when it starts. This amount should be enough in most cases. However, if your computer doesn't have enough free memory or you want to process very large documents with thousands of pages, you may need to modify the default memory settings. You can modify these settings by editing the chemcurator.conf (C:\Program Files\ChemAxon\ChemCurator\etc) file Xmx and Xms parameters.
default_options="--branding chemcurator -J-Dfile.encoding=UTF-8 -J-Xmx2048m -J-Xms256m -J-Dnetbeans.logger.console=true"Always create a copy of the configuration file before manually modifying it. Incorrect configuration settings may prevent the application from starting.