logD Plugin
This manual gives you a walk-through on how to use the logD Plugin:
Introduction
Compounds having ionizable groups exist in solution as a mixture of different ionic forms. The ionization of those groups, thus the ratio of the ionic forms depends on the pH. Since loP describes the hydrophobicity of one form only, the apparent logP value can be different. The logD represents the octanol-water coefficient of compounds at a given pH value.
Learn more about how the logD Plugin works.
The result window shows the pH:logD curves for each molecule drawn in the sketcher. The molecule images are shown in the legend. When clicking on an image, the corresponding molecule is displayed in the upper-left part of the panel.
The reference logD values originally shown can be reached by either clicking on the chart outside of the legend image areas or by selecting logD at reference pHs from the View menu.
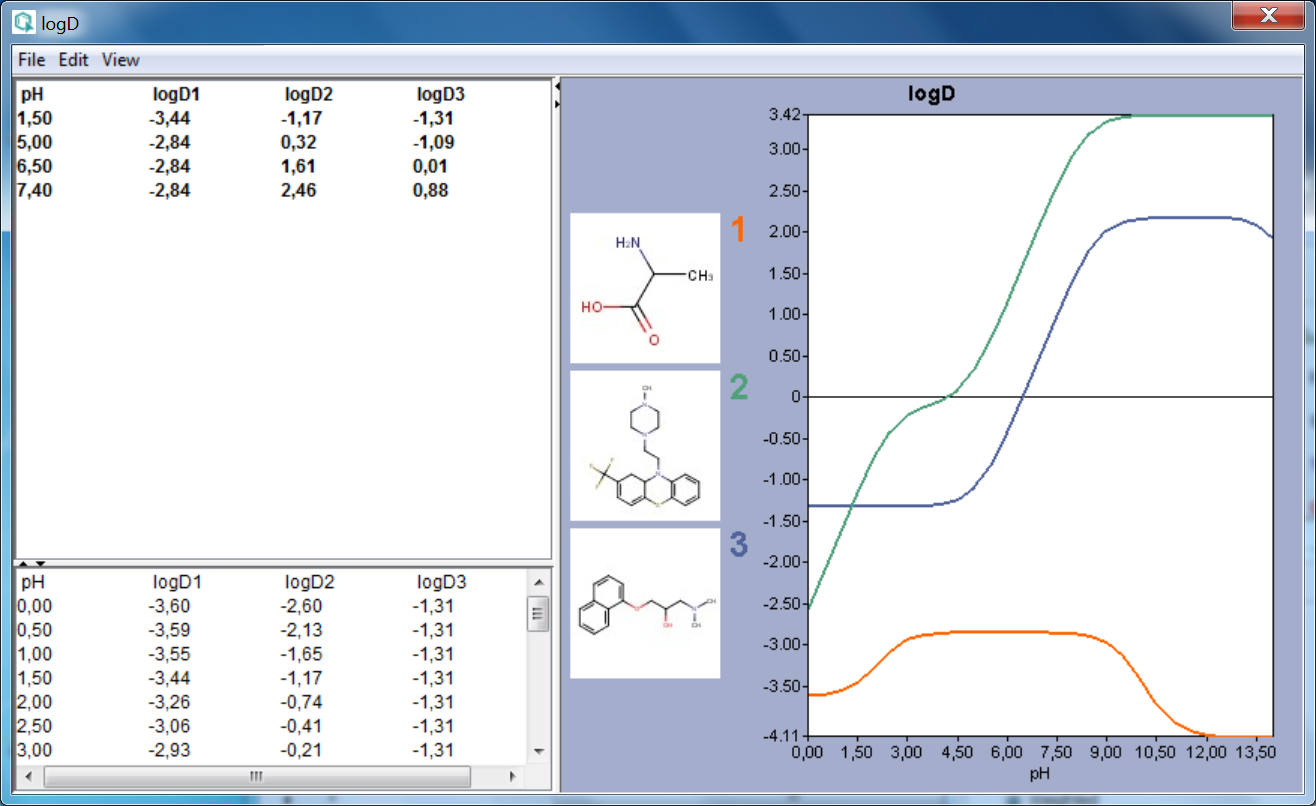
Fig. 1 logD result window showing the logD-pH curves for three molecules
Options
General Options
Different calculation (prediction) options can be set in the General Options tab of the logD Options window:
log P method
This option is for selecting the applied method for logP prediction. These can be:
-
Consensus: this method uses a consensus model built on the ChemAxon and Klopman et al. models and the PhysProp database.
-
ChemAxon: this method is based on ChemAxon's own logP model, which is based on the VG method (derived from Viswanadhan et al.). Read more about it here.
-
User defined: if a training set of structures and corresponding experimental logP values is available, it can be used as a database for logP calculations. See the manual page on creating such training sets.
-
Other options
These options are for refining the logD prediction.
-
LogP training ID: if the User defined method is selected, this dropdown list becomes active. All created training sets are listed here. Choose the one you want to apply for the calculation. Read more on creating a training set.
-
Electrolyte concentration
-
Cl- concentration: can be set between 0.1 and 0.25 mol/L.
-
Na+/ K+ concentration: can be set between 0.1 and 0.25 mol/L.
-
-
Use pKa correction library: a custom pKa training library for the compounds may be used. First, create a training set for your compunds, which then will appear in the dropdown list. If the option is checked, this list becomes active. Read more on creating a training set.
-
Consider tautomerization/resonance: in case of tautomer structures, all dominant tautomers at the given pH are taken into account during the logD calculation.
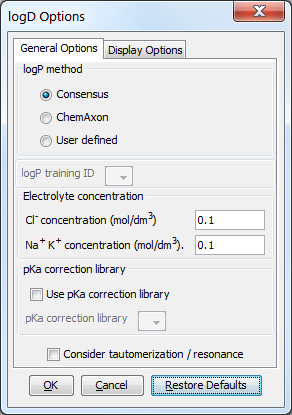
Fig. 2 General Options tab of the logD Options window
Display Options
Different display options can be set in the Display Options tab of the logD Options panel:
-
Decimal places: this sets the number of decimal places for the precision of the result value.
-
Chart: sets the parameters for the pH window in which the logD is calculated and displayed. The chart displays pH values starting from the lower limit incremented by the step size up until the upper limit. The results are given in table format as well.
-
Reference pH values: the logD at the given reference pH values are calculated with the predefined accuracy.
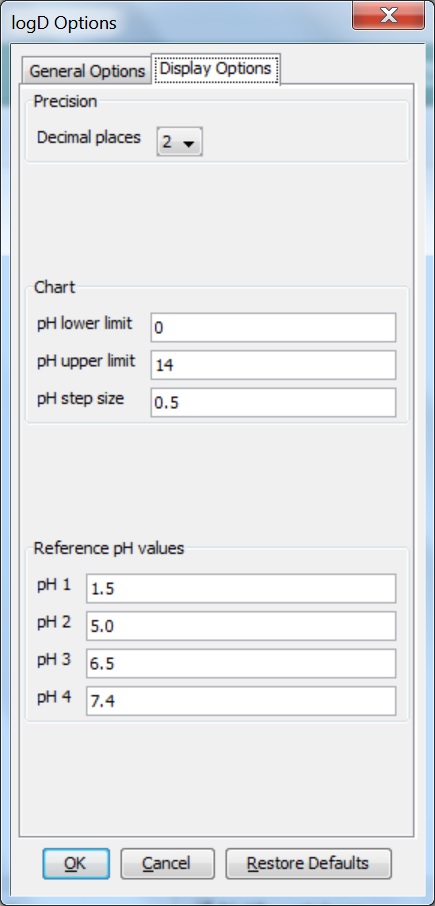
Fig. 3 Display Options tab of the logD Options window
References
-
Viswanadhan, V. N.; Ghose, A. K.; Revankar, G. R.; Robins, R. K., J. Chem. Inf. Comput. Sci., 1989, 29, 163-172; doi
-
Klopman, G.; Li, Ju-Yun.; Wang, S.; Dimayuga, M.: J.Chem.Inf.Comput.Sci., 1994, 34, 752; doi
-
PHYSPROP© database
-
Csizmadia, F; Tsantili-Kakoulidou, A.; Pander, I.; Darvas, F., J. Pharm. Sci., 1997, 86, 865-871; doi