MarvinView Table View Example
This example demonstrates the creation of scrollable molecule tables.
A mechanism by which molecules are loaded dynamically and cached automatically is also shown. This makes it possible that input files are supported with arbitrary size.
The result is shown with an example input below:
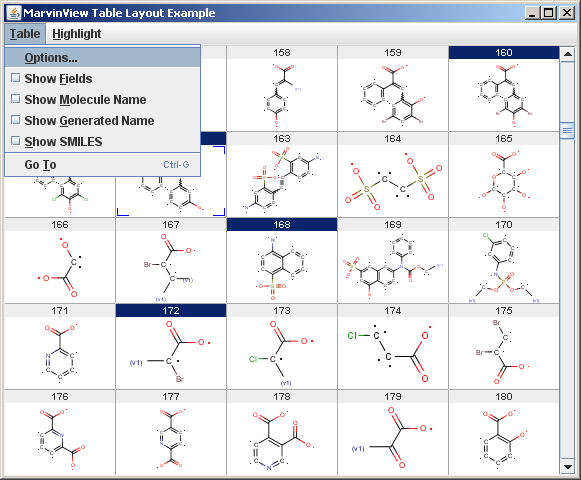
Specifying the input
To allow dynamic loading and caching the input of the viewer must be an implementation of MDocSource. You can either use an existing implementation like MolImporter or ArrayMDocSource, or you can create a custom implementation.
In the current example, the input is a molecule file hence it is convenient to use MolImporter:
static MDocSource createDocSource(String fname) { try { return new MolImporter(fname); } catch(FileNotFoundException ex) { System.err.println("File " + fname+" not found"); } catch(MolFormatException ex) { System.err.println("File " + fname + " is corrupted or not a structure file.\n" + ex.getMessage()); } catch (IOException e) { System.err.println("Error reading file " + fname+"\n"); } return null; }Creating the GUI
The creation of the GUI must be performed in the event dispatch thread.
// Swing components should be created, queried, and manipulated on // the event-dispatching thread according to Sun's recommendations. EventQueue.invokeLater( new Runnable() { public void run() { createAndShowGUI(docSource, fileName); } }); It consists of the following steps:
-
The creation of an MViewPane object:
MViewPane viewPane = new MViewPane(); viewPane.setBorderWidth(1); viewPane.setBackground(Color.LIGHT_GRAY); viewPane.setMolbg(Color.WHITE);
-
Adding the viewPane and a menubar to a JFrame:
JFrame win = new JFrame(); JMenuBar menubar = new JMenuBar(); win.setJMenuBar(menubar); win.setTitle("MarvinView Table Layout Example"); win.setDefaultCloseOperation(JFrame.EXIT_ON_CLOSE); win.getContentPane().setLayout(new GridLayout(1, 1)); win.getContentPane().add(viewPane); -
The creation of a TableSupport object is performed with the first MViewPane.getTableSupport() call:
TableSupport tableSupport = viewPane.getTableSupport();
-
Adding Table menu items using TableSupport.makeTableMenu:
JMenu menu; tableSupport.makeTableMenu(menu = new JMenu("Table")); menu.setMnemonic('t'); menubar.add(menu); -
Optional table settings using the TableOptions class:
TableOptions tblopts = tableSupport.getTableOptions(); // tblopts.setViewHandlerType(TableOptions.VH_GRIDBAG); tblopts.setMaxRows(5); tblopts.setMaxCols(5);
-
Making the window visible.
win.pack(); win.setLocationRelativeTo(null); win.setVisible(true);
-
Starting molecule table visualization using TableSupport.start(docSource, inputName), where docSource is the input document source (see the previous section) and inputName is a string describing the input, for example name of the input file — its function is to make error messages more specific.
tableSupport.start(docSource, inputName);
Changing the background of molecule ID fields
The Example menu contains a "Highlight" menu which demonstrates the highlighting of a record by changing the background color of its ID field.
The color of the ID field is changed using viewPane.setRecordIDBackground and setRecordIDForeground:
int selected = viewPane.getSelectedIndex(); viewPane.setRecordIDBackground(selected, SystemColor.textHighlight); viewPane.setRecordIDForeground(selected, SystemColor.textHighlightText);
Source code:ViewTable.java