Chemical Hashed Fingerprint
This manual gives you a walk-through on how to use Chemical Hashed Fingerprints:
Introduction
The chemical hashed fingerprint of a molecule is bit string (a sequence of 0 and 1 digits) that contains information on the structure. Chemical hashed fingerprints are mostly used in the following areas:
-
Chemical database handling: for full structure, substructure and similarity searching. It provides a rapid and effective screening, but in case of structural search it may generate false hits. For this reason the results have to be checked by a precise but slower atom-by-atom search.
-
Combinatorial chemistry: for the diversity/similarity analysis of compound libraries.
At both applications a proper configuration of the fingerprint is very important:
-
Structural search: the search becomes very slow if atom-by-atom searching receives too many false hits.
-
At similarity calculations the bitstrings should hold as much information as possible about the structure of the molecules.
ChemAxon provides the GenerateMD program for generating binary fingerprints that can be processed further. This program can also be applied to fine-tune fingerprint parameters for JChem.
Process of Fingerprint Generation in JChem and GenerateMD
The process of fingerprint generation goes as follows:
-
Up to a given a bond number all linear paths (linear patterns) consisting bonds and atoms of a structure are detected.
-
Branching points at the end of each linear pattern are also detected.
-
All cycles (cyclic patterns) are detected.
-
Using a proprietary hashing method, a given number of bits in the bit string are set for each pattern. It is possible, that the same bit is set by multiple patterns. This phenomenon is called bit collision. Few bit collisions in the fingerprint is tolerable, but too many may result in losing information in the fingerprint.
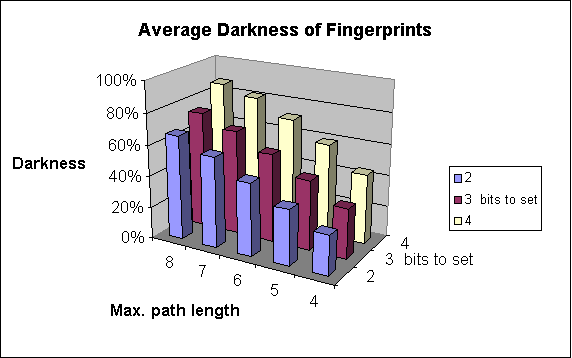
The figure below shows this process on an example.
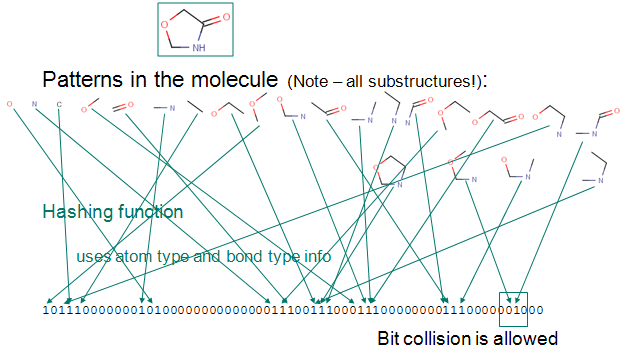
Chemical hashed fingerprint generation.
Fig. 1 Chemical Hashed Fingerprint generation process
Fingerprint parameters
Fingerprint length
The number of bits in the bit string.
Maximum pattern length
The maximum length of atoms in the linear paths that are considered during the fragmentation of the molecule. (The length of cyclic patterns is limited to a fixed ring size.)
Bits to be set for patterns
After detecting a pattern, some bits of the bit string are set to "1". The number of bits used to code patterns is constant.
Darkness of the fingerprint
The percentage of 1 digits in the bit string. We consider fingerprints with more ones "darker" than those with less ones.
Effect of Parameters
Effect of Increasing the Fingerprint Length
It increases
-
the capacity for storing information on molecules
-
the necessary disk space
-
the size of the structure cache
-
the time of structure import
-
the time of screening
In addition, it decreases fingerprint darkness, and therefore the probability of bit collisions also, which is beneficial.
A too long bit string may decrease the efficiency of information storage. We found that a length of 512 bits (64 bytes) worked well for small and huge databases as well. However, in similarity calculations longer fingerprints exhibit better performance in terms of selectivity (that is, distinguishing similar but not identical compounds). This is important in similarity based virtual screening as well as in similarity based clustering. In such applications 1024 bits usually provide better results.
Effect of Increasing Maximum Pattern Length
The effects are:
-
Longer and more patterns hold more information on the molecule.
-
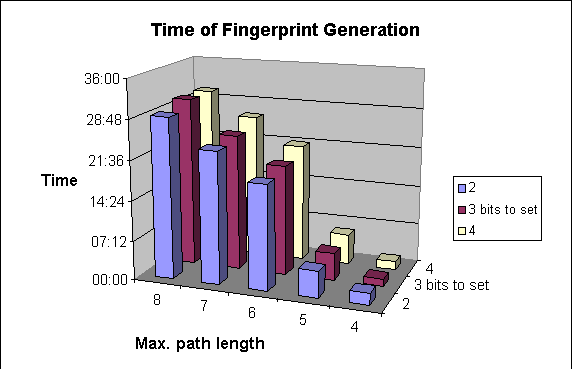
Due to the higher number of patterns to be explored, structure import will be slower.
-
Increases the number of patterns considered. As a result of that, the fingerprint darkness rises (up to a limit).
-
The number of bit collisions increase.
-
While the number of bit collisions is not too high, the stored information increases, which is beneficial for the efficiency of screening.
Substructure searching performs well with 5-6 long patterns. In similarity searching, however, longer patterns may be required, 7 is usually good value, and path longer than 8 seldom improve results. Also bear in mind that longer paths necessitates longer fingerprint to avoid too dark fingerprints.
Effect of Increasing the Number of Bits to Be Set for Patterns
The effects are:
-
The fingerprint darkness rises.
-
The coded information derived from a pattern increases.
-
The effect on darkness is similar to the case of the maximum pattern length: it also increases. And also, while the bit collision number is not too high, the stored information increases, which is beneficial for the efficiency of the screening.
Typical value is 2. Database pre-filtering for substructure searching does not require larger bit-count than 2, and this allows shorter fingerprints that is usually beneficial both in terms of storage space requirement and retrieval time. Though higher values could enable better separation of similar but not identical compounds thus leading to less frequent call of atom-by-atom matching in substructure searching, but only with the expense of doubled storage space and thus slower retrieval and more time consuming fingerprint comparison which is significantly more frequent procedure than the atom-by-atom searching.
Again, the situation is somewhat different in similarity searching, yet values higher than 2 rarely increase the amount of information represented by the fingerprint significantly (as the 3rd, 4th etc bits are more correlated with the other two, while 1st and 2nd are highly independent).
Optimizing Parameters For Search Efficiency
To choose optimal parameters for your compounds, running GenerateMD with the --stat option is recommended, or the use of JChem table or index statistics. (See more information in the JChem Manager command line usage (s command), at the Cartridge index statistics function and the Statistics tab at Instant JChem Schema editor.) These tools provide some practical information on the database (average/minimum/maximum "darkness", distribution, etc.).
Maximum darkness should not be higher than 80% (other sources/users say 2/3, ie. 67%). Otherwise, the information content of the individual fingerprint is decreased, and thus in similarity searching, for instance, similar though not identical compounds cannot be distinguished. Even a few too dark fingerprints also decrease screening efficiency at structure searching and consequently atom-by-atom search is unnecessarily often performed on the records with the dark fingerprints, even when target structures do not contain the given query structure.
The average darkness highly depends on the application and the particular data set (e.g. total diversity highly influences fingerprint darkness). In theory the information content is optimal at an average darkness of 50%, though in general, darkness should not exceed 40% to be on the safe side (to avoid frequent collisions).
The following statistics output shows an optimal fingerprint configuration, as generated by JChem table statistics function:
Statistics for table: APP.SCREENING_COLLECTION
--------------------
Row count: 58850
NULL SMILES count: 0
Average SMILES length: 40.09
Average compressed SMILES length: 20.34
Markush structure count: 0 (0.0%)
Fingerprint settings:
Length (bits): 768
Pattern length: 6
Bits set per pattern: 2
Min. CFP darkness: 4.03% cd_id: 26456
Max. CFP darkness: 69.66% cd_id: 20757
Avg. CFP darkness: 33.57%
Chemical Fingerpint distribution:
--------------------------------
0% - 10% : 0.6 %
10% - 20% : 8.54 %
20% - 30% : 28.8 %
30% - 40% : 34.92 %
40% - 50% : 21.42 %
50% - 60% : 5.27 %
60% - 70% : 0.42 %
70% - 80% : 0.0 %
80% - 90% : 0.0 %
90% - 100% : 0.0 %
The following graphs show the dependence of
-
average darkness of the fingerprints and
-
the time of the import of molecules
on two of the parameters of fingerprint generation. For these examples 64-byte-long bit strings were applied.