Biomolecules
Import biomolecules
You can import RNA, DNA and peptide sequences from the menu choosing File > Import As or through the Edit > Source panel. For more details please consult the documentation on sequence import.
In the second case you have to select the import mode ('Peptide Sequence', 'DNA Sequence', or 'RNA sequence') if it is not possible to decide whether the sequence belongs to a peptide or to a nucleic acid.
Peptides can be entered using their 1- or 3-letter amino acid codes
DNA nucleic acid sequences can be imported in four different formats:
-
ACGT,
-
A-C-G-T
-
dAdCdGdT or
-
dA-dC-dG-dT.
RNA nucleic acid import accepts sequences in two formats:
-
ACGU
-
A-C-G-U.
Display biomolecules
The sequences are displeayed in groupings of 20 residues.
DNA/RNA sequences are displayed with their 1-letter code on the canvas.
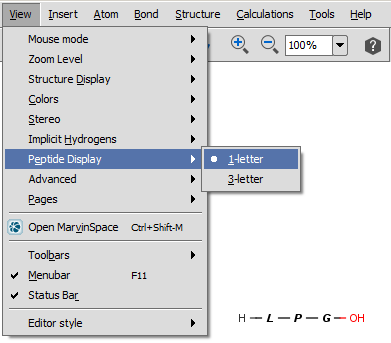
For peptides/proteins MarvinSkecth offers you the possibility to display them as their 1-letter or 3-letter codes selecting the View > Peptide Display menu item.
By setting the View > Colors > Peptide Bridges display option bonds crosslinking amino acids in different rows will be colored differently.
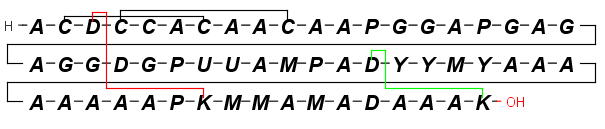
Draw bridges
You can easily draw peptidebridges and cycles in MarvinSketch. This way, you can better visualize bonds between amino acids, disulphide bridges, cyclic peptides, and lactam or lactone structures occuring frequently in peptides and proteins.
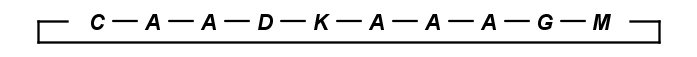
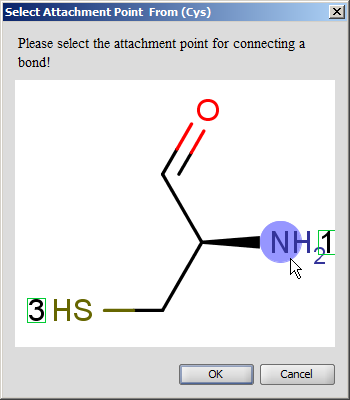
Select single bond tool and start to drag a bond from an amino acid: when you do this, the amino acids which have free attachment point(s) get highlighted. When you release the mouse, the new bond is established.
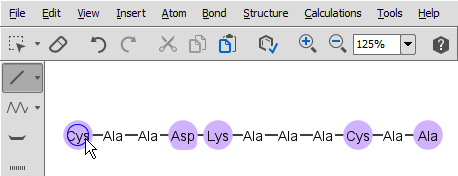
Do not forget to delete the explicit H and OH from the terminal amino acid chain if you would like to draw a head-to-tail cycle.
You cannot start a bond from an amino acid which does not have at least one free attachment point.
If there is more than one free attachment point on the starting and/or ending amino acid, you have to select the appropriate one from the pop-up dialog.