Typical Workflows
Standardizing chemical structures is a general component of drug discovery workflows. Representing compounds in clean and comparable formats is desired to ensure proper database design and promote database analysis.
In the following, two specific areas will be highlighted in which standardization of molecules comprises an essential step.
Compound registration
The compounds to be stored in a registration system usually go through validation, standardization and structure checking steps:
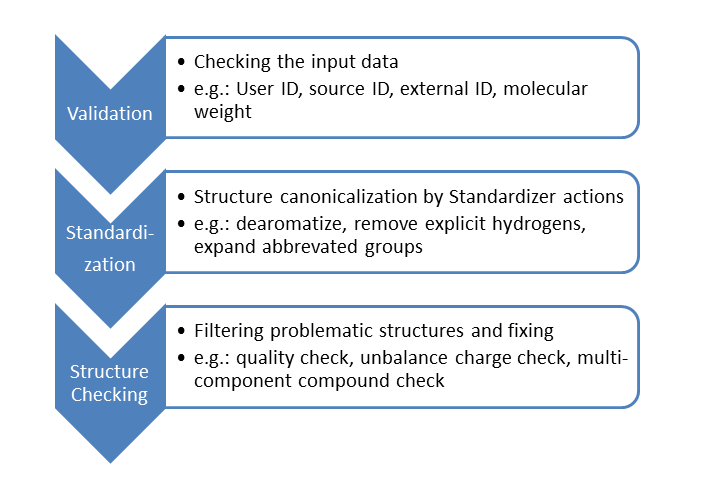
In the general validation step, the input data is thoroughly checked. If error is detected in any parameters, the compound cannot pass this step and manual amending is requred to fix the problem.
In order to assure high compound database quality, new compounds should enter the database in a previously specified uniform representation. Thus, structures are passed through the customized standardization actions of Standardizer to generate the requested chemical form.
Finally, structural problems are detected by structure checking. A structure labeled as problematic by a checker is either prevented from being registered or can be fixed automatically by the corresponding fixer.
Click here to learn about ChemAxon's Compound Registration system!
Structure-based virtual screening
Structure-based virtual screening using molecular docking algorithms is a widely used tool in the drug discovery process. In order to complete this task successfully, ligand databases should be preprocessed prior to virtual screening.
Large compound databases are usually filtered first to remove molecules with intrinsically nondruglike properties. The most common filter applied for this task is the Lipinski Rule of Five.
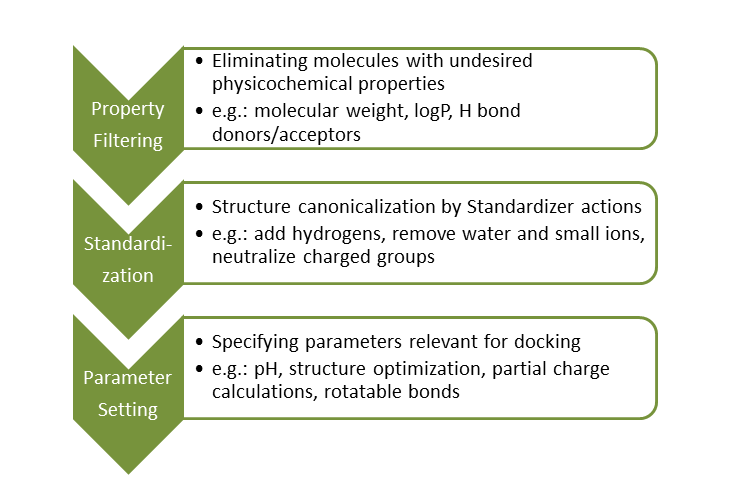
Molecules with appropriate physicochemical properties are then standardized to generate uniform molecular representations. This process eliminates database inconsistencies.
Lastly, docking-related parameters are set and ligands are transformed to a ready-to-dock format accordingly.