Extended Connectivity Fingerprint (ECFP)
This manual gives you a walk-through on how the ECFPs work:
Introduction
Extended-Connectivity Fingerprints (ECFPs) are circular topological fingerprints designed for molecular characterization, similarity searching, and structure-activity modeling. They are among the most popular similarity search tools in drug discovery and they are effectively used in a wide variety of applications.
ChemAxon provides the GenerateMD program for producing various molecular descriptors, including ECFPs, that can be processed further. This program can also be applied to fine-tune fingerprint parameters for JChem.
Applications
Circular fingerprints are effective and popular search tools, which have been successfully applied to a wide range of applications.
The initial application of ECFPs was in the area of high-throughput screening (HTS). In the evaluation of HTS results, ECFPs are widely used to analyze false positive and/or false negative hits. Furthermore, ECFPs are frequently applied in ligand-based virtual screening studies to distinguish between actives and inactives. Comprehensive studies revealed that these circular fingerprints are typically among the best performing search tools.
Various other areas of drug research related to similarity searching, including chemical clustering and compound library analysis, successfully utilize the rich information encoded in these fingerprints.
Beside similarity searching, ECFPs are well suited to the recognition of the presence or absence of particular substructures. Thus they are often used in QSAR and QSPR model building in the lead optimization phase, including the prediction of ADMET properties.
Properties of ECFPs
The main properties of ECFPs are the following:
-
They represent molecular structures by means of circular atom neighborhoods.
-
They can be very rapidly calculated.
-
Their features represent the presence of particular substructures.
-
They are not predefined and can represent a huge number of different molecular features (including stereochemical information).
-
They are designed to represent both the presence and the absence of functionality, since both are crucial for analyzing molecular activity.
-
Their generation method can be flexibly customized to produce various types of circular fingerprints for diverse applications.
Comparison to Path-Based Fingerprints
Path-based fingerprints, such as ChemAxon Chemical Fingerprint, were specifically designed and are widely used for pre-filtering in substructure searching. In contrast to this, ECFPs are not suitable for substructure searching, but they provide a rapid and highly effective screening method for full structure and similarity searching. Compared to path-based fingerprints, ECFPs typically provide more adequate results for similarity searching, which approximate the expectations of a medicinal chemist better.
Representation and Generation
ECFPs are not based on predefined substructural keys, but their features are generated in a molecule-directed manner. The ECFP generation process systematically records the neighborhood of each non-hydrogen atom into multiple circular layers up to a given diameter. These atom-centered substructural features are then mapped into integer codes using a hashing procedure. It is the set of the resulting identifiers that defines the extended-connectivity fingerprint.
Representations
ECFPs have two typical representations, both of which are supported by the ChemAxon implementation:
-
List of integer identifiers
The natural and accurate representation of ECFPs is by means of varying-length lists of integer identifiers. Each identifier represents a particular substructure, more precisely, a circular atom neighborhood, which is present in the molecule. The list of integer identifiers is sorted in ascending order.
These identifiers can also be interpreted as indexes of bits in a huge virtual bit string. Each position in this bit string accounts for the presence or absence of a specific substructural feature. Since this virtual bit string is extremely large and sparse, it is not stored explicitly, but the indexes of the 1 bits are recorded in a varying-length list.
In spite of this interpretation, the feature identifiers are stored as signed values due to technical reasons, that is, they can be either positive or negative.
By default, this integer list representation contains only one instance of each identifier. However, in particular applications, it could be beneficial to consider the frequency count of the ECFP features, that is, to record each identifier as many times as the represented feature occurs in the molecule. This variation of ECFP is often denoted as ECFC.
In our implementation, there is a configuration parameter that controls if the occurrence counts of the identifiers should be discarded (this is the default behavior) or kept (ECFC mode). -
Fixed-length bit string
Traditional representation of binary molecular fingerprints is by means of fixed-length bit strings. This representation can also be applied to ECFPs by "folding" the underlying virtual bit string into a much shorter bit string of specified length (e.g., 1024).
Compared to the identifier lists, this representation simplifies the comparison and similarity calculation of ECFPs and it could reduce the required storage space, especially for large molecules. On the other hand, the applied folding operation increases the likelihood of collision, that is, two (or more) different substructural features could be represented by the same bit position. As a result, a certain amount of information is usually lost, which worsens both the quality and interpretability of this representation.
Note that the fixed-length binary representation can be derived from the identifier list representation, but the opposite transformation is not possible. In other words, the fixed-length bit string representation can be viewed as a "lossy" compression of the identifier list (or the underlying virtual bit string).
Generating ECFPs in different ways
The following tools are available for generaing ECFPs:
-
GenerateMD command line tool
Using the GenerateMD command line application, the -D option selects the identifier list output, and the -2 option selects the fixed-length bit string output, e.g.generatemd c input.smiles -k ECFP -c ecfp_config.xml -2 -
DescriptorGenerator API
Using the DescriptorGenerator class, the functions getAsString() and getAsIntArray() give back the identifier list representation, while getAsBitSet() gives back the fixed-length bit string representation.
-
ECFP API
Using the ECFP class, the functions toString(), toIntArray(), and toIdentiferSet() give back the identifier list representation, while toBinaryString() and toBitSet() give back the fixed-length bit string representation.
The Generation Process
The fingerprint generation process is as follows:
-
Initial assignment of atom identifiers
The ECFP generation process begins with the assignment of an initial integer identifier to each non-hydrogen atom of the input molecule. This identifier captures some local information about the corresponding atom in such a way that various atom properties (e.g., atomic number, connection count, etc.) are packed into a single integer value using a hash function.
The set of considered atom properties is an important configuration parameter of ECFPs, which can be fully customized (see later). -
Iterative updating of identifiers
After that, a number of iterations are performed to combine the initial atom identifiers with identifiers of neighboring atoms until a specified diameter is reached. Each iteration captures larger and larger circular neighborhoods around each atom, which are then encoded into single integer values using a suitable hashing method and these identifiers are collected into a list.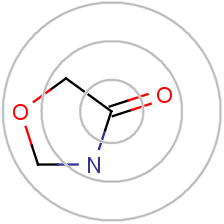
Fig. 1. Illustration of the effect of iterative updating for a selected atom in a sample moleculeThis iterative updating process is based on the well-known Morgan algorithm.
-
Duplication removal
If the identifier counts should be kept, then this step is modified to store each integer identifier as many times as the corresponding substructural feature occurs in the molecule. The final step of the generation process is the removal of multiple identifier representations of equivalent atom neighborhoods. Two neighborhoods are considered to be equivalent if they contain exactly the same set of bonds or their hashed integer identifiers are the same.
The following figures illustrate the whole ECFP generation process, including the derivation of the fixed length bit string from the identifier list representation.
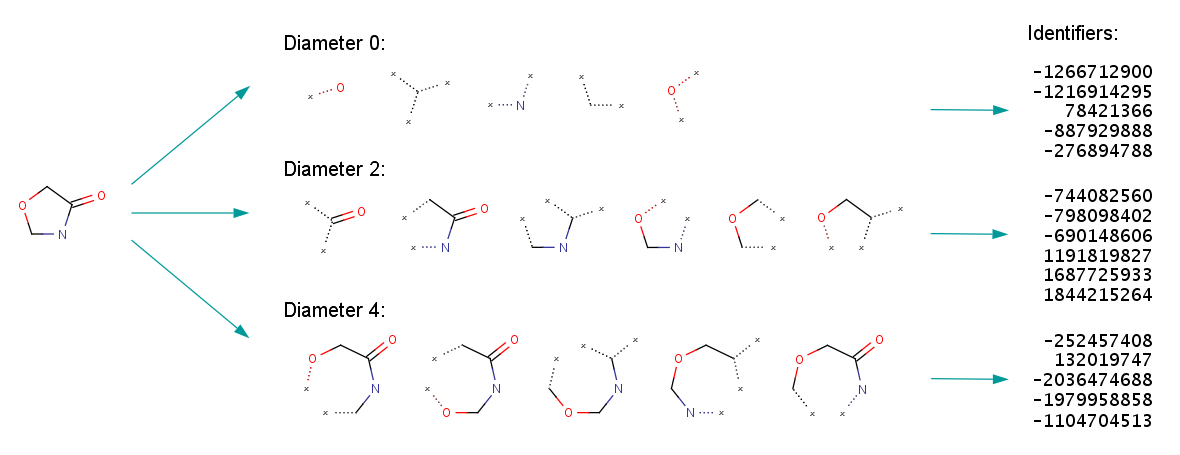
Fig. 2. ECFP generation process
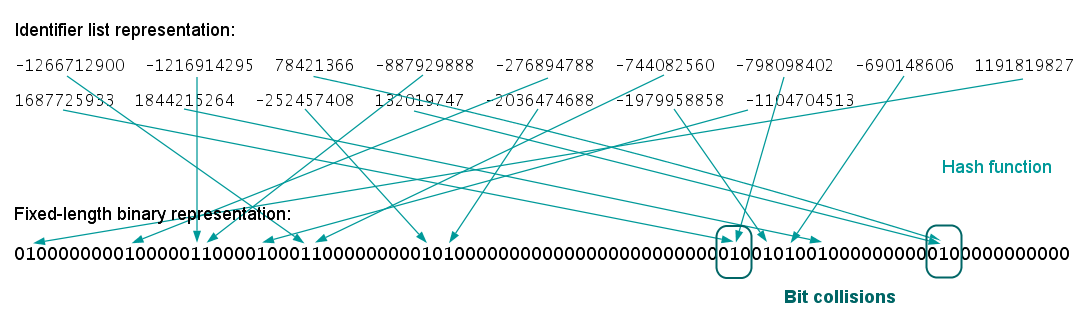
Fig. 3. Generation of the fixed-length bit string ("folding")
ECFP configuration
ECFPs provide a rich set of configuration parameters. They can be specified in an XML file, just like the configuration of other molecular descriptors.
Main parameters
The three main parameters of ECFPs are maximum diameter, fingerprint length, and identifier counts:
-
Diameter
This parameter specifies the maximum diameter of the circular neighborhoods considered for each atom. The default diameter is 4. This is a dominant parameter of ECFPs, which controls the number and the maximum size of considered atom neighborhoods, thus it controls the length of the identifier list representation, as well as the number of "1" bits in the fixed length bit string representation. (More information about these two representations can be found above.)
ECFPs are usually distinguished by this parameter. For example, ECFP_4 denotes that the maximum diameter is set to 4, while ECFP_6 means diameter 6.
The appropriate value of the maximum diameter depends on the desired application. According to Rogers and Hahn, diameter 4 is typically sufficient for similarity searching and clustering, while activity learning methods often benefit from the greater structural detail available by using larger limit, for example 6 or 8. -
Length
This parameter specifies the length of the bit string representation. The default length is 1024.
Larger length decreases the likelihood of bit collision, and therefore decreases the information loss. However, handling larger fingerprints require more computation time and storage space. -
Counts
This parameter controls whether the generated integer identifiers are stored with occurrence counts or each identifier is kept only once independently of the number of the corresponding substructural features in the input molecule. The default option is "No", that is, each identifier is stored only once.
The first two parameters play similar role to the maximum pattern length and fingerprint length parameters of ChemAxon Chemical Hashed Fingerprint. They have similar effects on the information content, the generation time, and the required storage space of the fingerprints.
These main parameters of ECFPs can be specified as attributes of the <Parameters/> tag in the XML configuration file, e.g.
<Parameters Length="512" Diameter="2" Counts="Yes"/>Atom Properties
The set of atom properties encoded in the atom identifiers is an important configuration parameter of ECFPs, which determines their characteristic. Different sets of properties result in different types of circular fingerprints targeting various applications.
The default ECFP configuration supports typical use cases, especially similarity searching based on highly specific substructural information. For each atom, the following properties are considered by default:
-
the atomic number;
-
the number of "heavy" (non-hydrogen) neighbor atoms;
-
the number of attached hydrogens (both implicit and explicit);
-
the formal charge;
-
an additional property that indicates whether the atom is part of at least one ring.
If we would like to use other identifier configuration, we can make a selection of a few built-in properties and a wide variety of custom properties defined by Chemical Terms.
Examples
-
The default identifier configuration can be described as follows:
<IdentifierConfiguration><Property Name="AtomicNumber" Value="1"/><Property Name="HeavyNeighborCount" Value="1"/><Property Name="HCount" Value="1"/><Property Name="FormalCharge" Value="1"/><Property Name="IsRingAtom" Value="1"/></IdentifierConfiguration>The built-in properties are identified by their names. The Value attribute specifies if the atom property is actually used (1) or not (0). It facilitates enabling and disabling atom properties without the permanent removal of the unused properties from the file. The default configuration file contains all built-in properties, but only the above five are enabled. The order of the properties in the XML file is arbitrary, and it has no effect on the generated fingerprints.
-
A configuration using other built-in properties:
<IdentifierConfiguration><Property Name="MassNumber" Value="1"/><Property Name="ConnectionCount" Value="1"/><Property Name="HasAromaticBond" Value="1"/><Property Name="IsTerminalAtom" Value="1"/><Property Name="IsStereoAtom" Value="1"/></IdentifierConfiguration>
-
A configuration using custom properties defined by Chemical Terms:
<IdentifierConfiguration><Property Name="MyAtomicNumber" Value="1">atno()</Property><Property Name="MyConnectionCount" Value="1">connections()</Property><Property Name="PosCharge" Value="1"><![CDATA[ charge() > 0.4 ]]></Property><Property Name="NegCharge" Value="1"><![CDATA[ charge() < -0.3 ]]></Property></IdentifierConfiguration>The first two properties are equivalent to the corresponding built-in properties. They are expressed using Chemical Terms only for illustration purposes. Note that the built-in properties are typically computed faster, thus they should be preferred if it is possible.
On the other hand, the partial charge can only be expressed using Chemical Terms. The latter two properties are practical examples for defining custom properties based on the partial atomic charge.
Non-integer property values
For technical reasons, all property values are converted to integers. The true/false (boolean) values are converted to 1 and 0, respectively, while the floating point values are simply truncated to integers, that is, their fractional parts are discarded (not rounded). This rough truncation could eliminate differences that we would like to consider in the ECFP computation. In such cases, we can multiply the original floating point value by an appropriate factor.
For example, if we would like to consider the partial charge up to one decimal digit, then we can use
<Property Name="Charge" Value="1"><![CDATA[ charge() * 10 ]]></Property>instead of
<Property Name="Charge" Value="1"><![CDATA[ charge() ]]></Property>This latter property would not distinguish charge values 0.16, 0.32, 0.38, -0.55 etc. (all of them would be converted to 0), while the first property would convert them to 1, 3, 3, and -5, respectively.
Functional-Class Fingerprints (FCFPs)
The default identifier configuration of ECFP captures highly specific atomic information enabling the representation of a large set of precisely defined structural features. In some applications, however, different kinds of abstraction may be desirable. For example, a chlorine or a bromine substituent on a ring may be functionally equivalent but would be distinguished by standard ECFP. Another example that opens a new direction in the application of circular fingerprints is the pharmacophore identification of atoms which transforms ECFPs to topological pharmacophore fingerprints.
Those variants of ECFPs that applies such generalizations and have focus on the functional roles of the atoms instead of full specificity are calledFunctional-Class Fingerprints (FCFPs).
ChemAxon's ECFP implementation also supports FCFPs by custom identifier configurations, since the generation of the initial atom identifiers is the only difference between ECFPs and FCFPs. For example, the following configuration defines a functional-class circular fingerprint:
<IdentifierConfiguration> <Property Name="HydrogenBondAcceptor" Value="1">acceptor()</Property> <Property Name="HydrogenBondDonor" Value="1">donor()</Property> <Property Name="Aromatic" Value="1">arom()</Property> <Property Name="Charge" Value="1"><![CDATA[ charge() * 10 ]]></Property> </IdentifierConfiguration>The full configuration file can be found here.
Note that abstraction of FCFPs typically results in smaller set of features than ECFPs for a given molecule, because different substructures may be functionally equivalent and hence the same identifier is generated for them.
Feature Retrieval
JChem also provides a lookup service for the features encoded in ECFP fingerprints. As we have discussed above, ECFPs are represented either as lists of integer identifiers or as fixed-length bit strings, in which the identifiers and bit positions account for particular substructural features of the input molecule. The ECFPFeatureLookup class serves the retrieval of these circular atom neighborhoods corresponding to a given integer identifier or bit position.
In some applications of ECFPs, the fingerprints of certain compounds are compared in order to identify particular feature identifiers or bit positions that seem to be important in characterizing or differentiating the molecules. In these cases, one may be interested in obtaining the actual substructures that are represented by the considered identifiers or bits.
For example, given an integer identifier id that is present in the ECFP fingerprint of a molecule mol, the corresponding substructures can be retrieved in SMARTS format with a code like this:
ECFPFeatureLookup lookup = new ECFPFeatureLookup(); lookup.processMolecule(mol); for (ECFPFeature f : lookup.getFeaturesFromIdentifier(id)) { System.out.println(f.getSubstructure().toFormat("SMARTS")); }For more information, see the API documentation of ECFPFeatureLookup .
References
-
Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50(5): 742-754.
-
Hu, Y.; Lounkine, E.; Bajorath, J. Improving the Search Performance of Extended Connectivity Fingerprints through Activity-Oriented Feature Filtering and Application of a Bit-Density-Dependent Similarity Function. ChemMedChem 2009, 4: 540-548.
-
Glen, R. C.; Bender, A.; Arnby, C. H.; Carlsson, L.; Boyer, S.; Smith, J. Circular fingerprints: Flexible molecular descriptors with applications from physical chemistry to ADME. IDrugs 2006, 9(3): 199-204.
-
Morgan, H. L. The Generation of a Unique Machine Description for Chemical Structures - A Technique Developed at Chemical Abstracts Service. J. Chem. Doc. 1965, 5: 107-112.