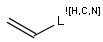Appendix
Differences in matching Daylight and MDL formats
We pursue compatibility with both MDL and Daylight structure searches. However, some query features have different meanings in the two systems. For this reason the interpretation of some query features depends on the query input format. Queries of type SMILES, SMARTS, cxsmiles and cxsmarts will be matched the Daylight way and all others the MDL way.
The affected query features and their different matchings are detailed below.
ANY and not list atoms
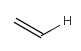
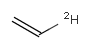
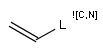
In the MDL terminology, ANY atoms never match hydrogens. This also excludes plain H, deuterium, charged H, etc. However, at Daylight ANY matches isotopic and charged H, but not plain Hydrogens.
In case of not list atoms, if H (or #1) does not appear in the excluded list, Daylight terminology behaves similarly as above: accept isotopic and charged H only. On the other hand, MDL never accepts Hydrogens for not lists. Here we chose not to comply with the MDL behavior even in the case of MDL format input to avoid misinterpretation. So in case of MDL format query all Hydrogens match to not lists. (Certainly if H atom type is included in the not list it will NOT match to H.) See examples below.
Table 1.
|
Query |
Targets |
||
|
|
|
|
|
|
MDL Query (molfile) |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Daylight Query (SMARTS) |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
H query property
In the MDL terminology, query property H <number> means at least <number> Hydrogens in excess explicitly drawn on the query. H0 is a special case which means no Hydrogens in excess the explicitly drawn. On the other hand, at Daylight H <number> means a total of <number> Hydrogens. (Explicit and implicit.)
Table 2.
|
Query |
Targets |
||
|
|
|
|
|
|
MDL Query (molfile) |
|||
|
|
|
|
|
|
|
|
|
|
|
Daylight Query (SMARTS) |
|||
|
|
|
|
|
|
|
|
|
|
Double bond stereo matching mode
This is related to cis-trans isomerism of double bonds. As described above, there is a search option to control this: setDoubleBondStereoMatchingMode(), defaulted to DBS_MARKED. When DBS_MARKED option is set, cis/trans is only considered at marked double bonds. (An MDL query feature, also called stereo care flag. It is depicted as a square over the double bond.) However, the Daylight terminology lacks marked double bonds, they use directional bonds: / and \ instead. In order to correctly evaluate stereo SMARTS queries using the default search in case of Daylight format queries, the DBS_MARKED option considers directional bonds. (Please note that there is no special depiction of these SMARTS stereo bonds in Marvin, however the non-stereo double bonds like CC=CC are depicted by a wiggly bond ligand.)
'D' and 's' features
The SMARTS feature 'D' (degree) in Daylight implementation by default does not follow its description ("explicit connections"): ignores explicit H connections (but counts explicit H isotopes). This is the same semantics as the MDL feature 's' ("substitution count") offers, so in searches the two features have the same meaning.
SMARTS feature matrix
Supported SMARTS features
Table 3.
|
SMARTS notation |
Description |
|
cC |
Aromatic/Aliphatic atoms |
|
* |
Any atom |
|
a |
Aromatic |
|
A |
Aliphatic |
|
<n> |
Isotope |
|
H<n> |
Total H count |
|
R<n> |
Ring membership |
|
r<n> |
Ring size |
|
v<n> |
Valence |
|
X<n> |
Connectivity |
|
+/- |
Charge |
|
#n |
Atomic number |
|
@, @@ |
Tetrahedral chirality |
|
@? |
Chiral or unspec |
|
bond types |
|
[#6,#7,#8,#9] |
Atom list |
|
[!#6!#14!#32!#50!#82] |
Atom not list |
|
[C:1] |
Map |
|
O>>O |
Reaction SMARTS |
|
(C.C) |
Component level grouping |
|
/? ? |
directional bond or unspecified |
|
D<n> |
Degree |
|
h<n> |
Implicit H-count |
|
@ |
Any ring bond |
|
! & ; , |
General logical expressions within atom and bond descriptions. |
|
$() |
Recursive SMARTS |
NOT YET supported SMARTS features
Table 4.
|
SMARTS notation |
Description |
|
@<c><n> |
Chirality class |
|
@<c><n>? |
Chirality class or unspec |
Molfile (MDL) query feature matrix
Supported Molfile(MDL) query features
Table 5.
|
Generic atoms: hetero(Q), Any(A) |
|
Atom list |
|
Atom not list |
|
No implicit hydrogens |
|
Valence(v<n>) |
|
Charge |
|
Isotope |
|
Radical |
|
Atom to atom map(reactions) |
|
Chiral atoms |
|
Chiral flag of molecules |
|
Enhanced stereo representation(ABS AND<n> OR<n>) |
|
Bond types: single, double, triple, aromatic, double cis or trans, single or double, single or aromatic, double or aromatic, any |
|
Stereo bond types: single up, single down, single up or down |
|
Double bond stereo care flag |
|
Reactions: starting materials, products |
|
Reaction stereo: inversion, retention |
|
Reacting center |
|
Atom alias |
|
Pseudo atoms |
|
LP atom type |
|
R-group queries: up to two connections per R-group |
|
R-logic: occurrence range, restH, if-then |
|
S-groups: Super atom (abbreviated group), multiple group, mixture, component, formulation |
|
Bond topology: in ring, in chain, none |
|
Unsaturated atom |
|
Ring bond count(RB) |
|
Substitution count |
|
Link atom |
|
Polymer and attached data S-group types |
NOT YET supported Molfile(MDL) features
Table 6.
|
3D special features |
|
Exact change flag (reaction) |
|
Beilstein generics |