HLB Predictor
This manual gives you a walk-through on how to use the HLB Predictor:
Introduction
The hydrophilic-lipophilic balance number (HLB number) measures the degree of a molecule being hydrophilic or lipophilic. This number is calculated based on identifying various hydrophil and liphophil regions in the molecule. This number is a commonly used descriptor in any workflow in which lipid based delivery can be an option. (e.g. lipid-based drug delivery, cosmetics)
The HLB Predictor is a general tool for predicting the HLB number. It can predict the HLB number based on the Davies or the Griffin method.
The accuracy of the HLB Predictor was measured using a test set of 26 molecules with available measured HLB values. The experimental molecules and their measured HLB values were taken from the reference articles.
The plot below shows how the predictor performed on this test set. The calculated R2 is 0.79.
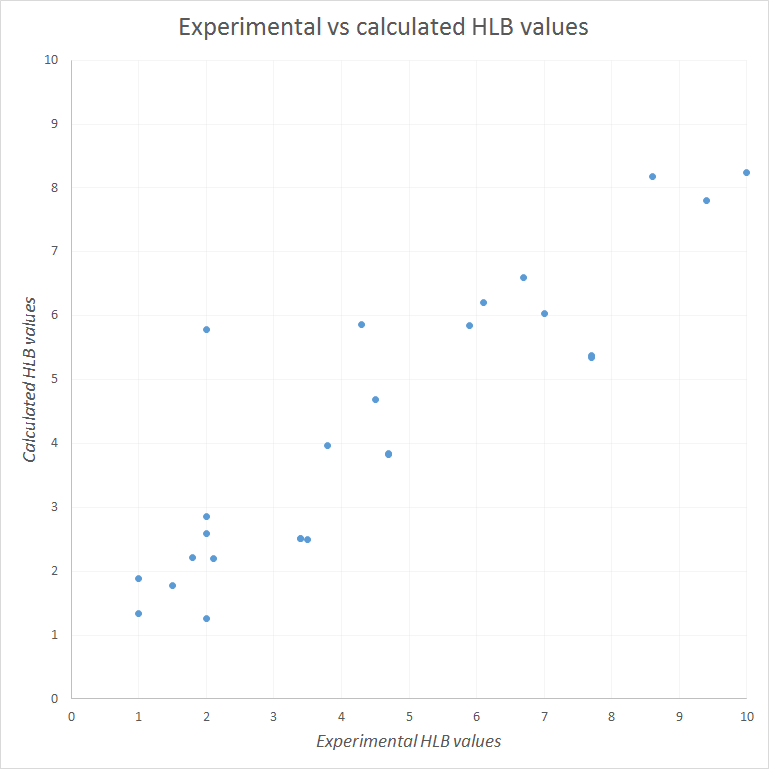
Fig. 1 Plot showing the accuracy of the HLB Predictor on a test set of 26 molecules
Usage
The HLB Predictor is integrated in various ChemAxon products.
MarvinSketch
The HLB Predictor is available as a plugin in MarvinSketch. It can be found under Calculations > Partitioning > HLB. The plugin has the following methods for calculation:
-
ChemAxon: this is a consensus method based on the other two methods with optimal weights (default)
-
Davies: this is an extended version of the Davies method
-
Griffin: this is an extended version of the Griffin method
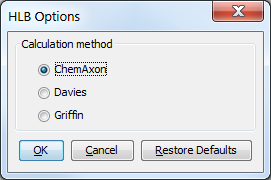
Fig. 2 The HLB Options window showing the available calculation methods
The calculation result appears in a separate pop-up window with the input molecule.
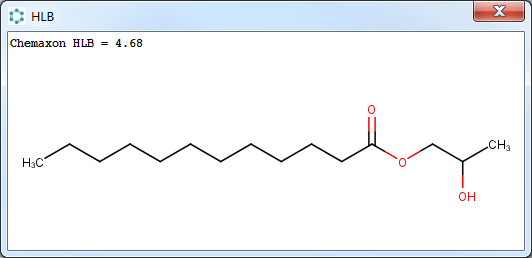
Fig. 3 The result window with the predicted HLB value and the input molecule
cxcalc
To use the HLB Predictor from the cxcalc command line tool, use the following syntax:
cxcalc [general options] [input files/strings] hlb [hlb options] [input files/strings]
The following options are available for the HLB predictor:
-h, --help this help message-p, --precision <floating point precision as number of fractional digits: 0-8 or inf> (default: 2)-m, --method [chemaxon|davies|griffin] (default: chemaxon)
Here are some examples of how to use the HLB Predictor via cxcalc:
-
Calculating the HLB number for an input molecule stored in SD format using the default method:
cxcalc hlb test.sdf -
Calculating the HLB number for an input molecule stored in SMILES using the Griffin method with 3 digit precision:
cxcalc hlb -m griffin -p 3 test.smiles
Chemical Terms
The HLB prediction functionality is also available in the Chemical Terms language via the hlb() function. The function has one parameter, the name of the method. If no parameter is given, the calculation runs with the default method.
Here is an example of how to calculate the HLB number via the Chemical Terms language:
evaluate -e "hlb('davies')" test.sdf
KNIME
The HLB Predictor is also available as a ChemAxon KNIME node. You can see the KNIME node Options panel with the same options as in MarvinSketch below.
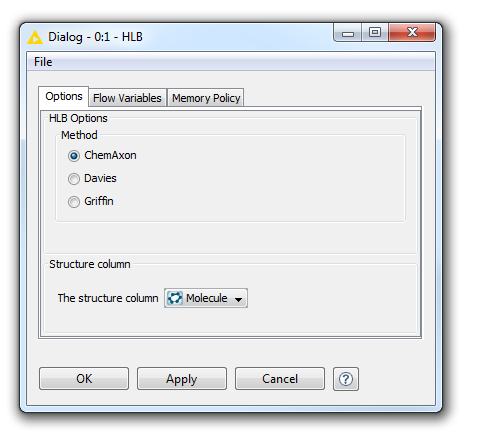
Fig. 4 HLB Options panel in KNIME
References
-
Davies, J.T. A quantitative kinetic theory of emulsion type, I. Physical chemistry of the emulsifying agent Gas/Liquid and Liquid/Liquid Interface (Proceedings of the International Congress of Surface Activity), 1957, 426–38
-
Griffin, W. C. Calculation of HLB Values of Non-Ionic Surfactants , Journal of the Society of Cosmetic Chemists, 1954, 5 (4), 249-56