Hit display/coloring
Contents
Introduction
The visualization of the search results is a very important feature of the JChem products.
The hitlists - especially in case of Markush targets - contain complex, not easily transparent chemical structures. Our hit display/coloring tool helps to elucidate the hit structures, makes easier to notice the conformity of the hit and the query structures. It uses two main methods: alignment and coloring. The display of Markush hits and result of similarity searches have further special options.
Alignment
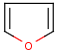
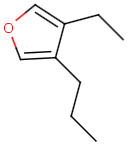
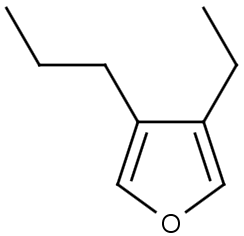
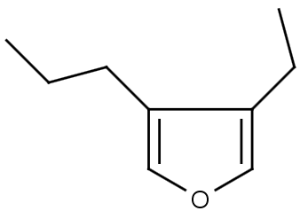
The alignment of the hit structure means that the position of the hit structure is aligned to that of the query structure.
-
By default the hit structure's position on the screen is the same as that of the target structure.
-
The hit structure's position on the screen is aligned to the query structure when alignment is switched on, that is the hit structure is rotated till its part corresponding to the query gets the same position as the query structure has.
-
The hit structure's position on the screen is partially aligned to the query structure when partial alignment is switched on. Partial clean is template based clean.
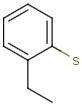
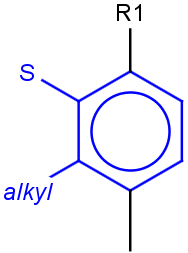
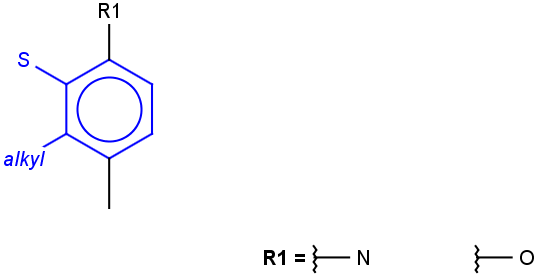
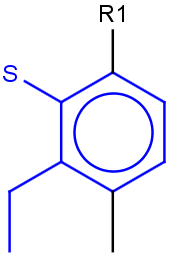
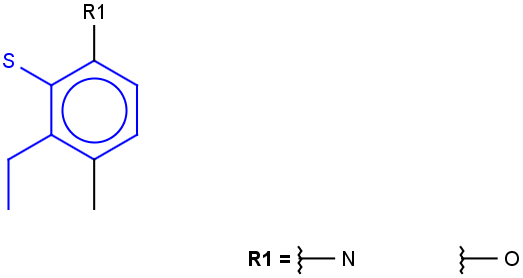
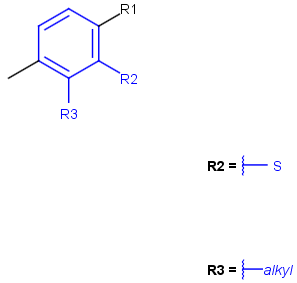
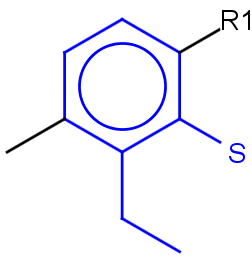
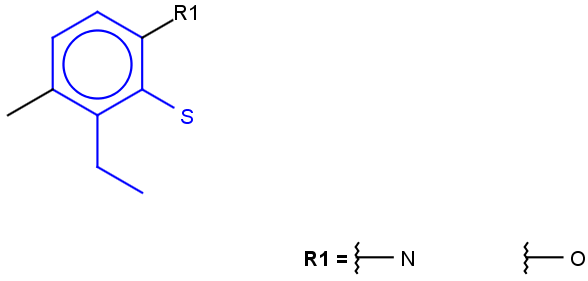
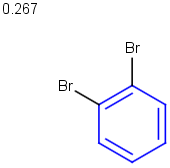
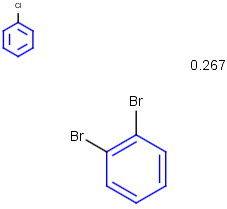
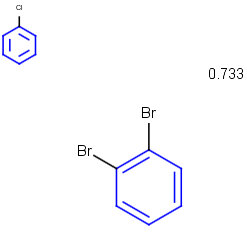
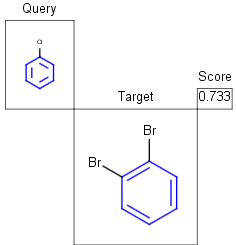
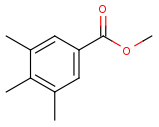
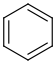
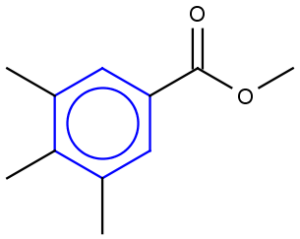
See examples in Table 1.
The detailed description of the hit alignment options on different search platforms can be found here.
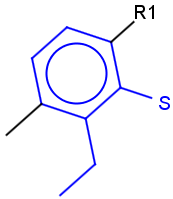
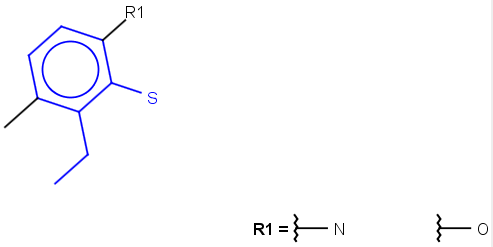
Table 1.
|
Query |
Target |
Hit alignment |
||
|
No/Off |
Rotate/On |
Partial clean |
||
|
|
|
|
|
|
Coloring
The visualization of the query structure as substructure in the hit can be improved by using the coloring features.
The color of the hit substructure, as well as the color of the non-hit substructure and the color of the homology groups within the hit structure can be customized.
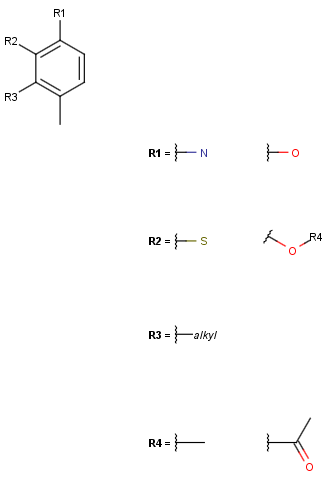
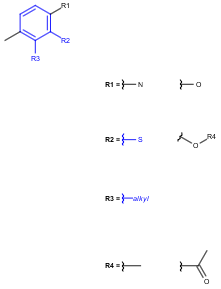
See examples in Table 2.
The detailed description of coloring options on different search platforms can be found here.
Table 2.
|
Query |
Target |
Hit coloring |
|
|
|
|
Hit color: red |
Hit color: red/Non-hit color: green |
|
|
|
|
Hit homology color: red |
Display of Markush hits
Beside the alignment and coloring option you can apply further display options to enhance the visualization of Markush hits.
One of these options is the "Markush display mode" option (see its detailed description on different platforms here);
the other option is the "Remove unused definitions" options (see its detailed description on different platforms here).
Markush dispaly mode options are the following:
-
Original
-
Markush reduction to the hit
-
Markush reduction and expansion of homology groups
Remove unused definitions option can be switched on/off.
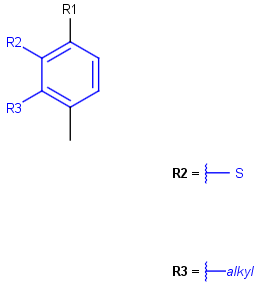
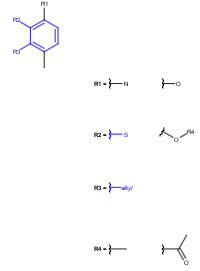
See examples in Table 3.
Table 3.
|
Query |
Target |
|
|
|
|
Alignment: Off |
||
|
Markush display mode |
Remove unused definitions |
|
|
Yes |
No |
|
|
Original |
|
|
|
Markush reduction to the hit |
|
|
|
Markush reduction and expansion of homology groups |
|
|
|
Alignment: Rotate |
||
|
Markush display mode |
Remove unused definitions |
|
|
Yes |
No |
|
|
Original |
|
|
|
Markush reduction to the hit |
|
|
|
Markush reduction and expansion of homology groups |
|
|
|
Alignment: Partial clean |
||
|
Markush display mode |
Remove unused definitions |
|
|
Yes |
No |
|
|
Original |
|
|
|
Markush reduction to the hit |
|
|
|
Markush reduction and expansion of homology groups |
|
|
Hit display in similarity search
The following hit display options are available in similarity search:
-
Display or not display (default) query structure;
-
Display similarity score (default), display dissimilariry score, display none of them;
-
Display or not display (default) labels and boxes. See examples in Table 4 .
The detailed description of hit display options in similarity search on different platforms can be found here.
Table 4.
|
Query |
Target |
|
|
|
|
Hit |
|
|
Default |
|
|
Display query structure |
|
|
Display query structure Display dissimilarity score |
|
|
Display query structure Display dissimilarity score Display labels and boxes |
|
Hit display in superstructure search
In case of superstructure search the target structure is highlighted as substructure within the hit structure. Alignment options (rotate and partial clean) are not applicable.
See example in Table 5.
Table 5.
|
Query |
Target |
Hit |
|
|
|
|