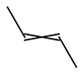
Stereoisomers around double bonds
Cis/Trans stereoisomers
The cis/trans information for ligands of the double bond can be calculated by scpecifying a ligand (a1) of the double bond connecting to one end (a2) of the double bond and giving another ligand (a4) of the double bond connecting to the other end (a3) of the double bond.
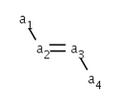
The possible values:
-
0: if no stereo information given or the double bond doesn't have enough ligands to be stereochemically active
-
CIS: if the ligands are on the same side of the double bond
-
TRANS: if the ligands are on the opposite side of the double bond
-
CIS|TRANS: if the ligands can be both CIS and TRANS arrangement
|
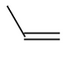
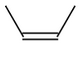
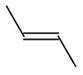
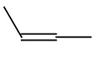
Molecule |
|
|
|
|
|
|
cis/trans value |
0 |
CIS |
TRANS |
CIS|TRANS |
CIS|TRANS |
An additional flag is used in conjunction with CIS or TRANS information in case of query molecules:
-
CTUNSPEC : the ligands are in the specified (CIS or TRANS) configuration or unspecified
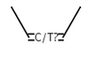
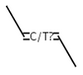
CIS or unspecified double bond
TRANS or unspecified double bond
Depending on the molecule's dimension the cis/trans information corresponding to the ligands of the double bond is stored in the bond or calculated from the coordinates.
It is optional to check atom equivalences using graph invariants for the ligands of the double bond during stereo calculation. If graph invariants are equal on one endpoint of the double bond then the two ligands connecting to this node are equivalent and so the double bond does not exhibit cis/trans isomerism.
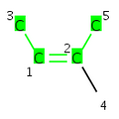
Double bond stereo information of the atoms 3-1=2-5
-
if graph invariants not checked on the ligands the result is CIS
-
if graph invariants checked on the ligands the result is 0
/* * Copyright (c) 1998-2014 ChemAxon Ltd. All Rights Reserved. */ import java.io.IOException; import chemaxon.struc.Molecule; import chemaxon.struc.MolBond; import chemaxon.struc.CNode; import chemaxon.struc.StereoConstants; import chemaxon.formats.MolImporter; /** * Example to get double bond stereo information of double bonds. * Usage: * java CisTransExample filename * * @version 5.1 04/24/2008 * @since Marvin 5.1 * @author Andras Volford */ public class CisTransExample { /** * Main method. * @param args command line arguments (filename) */ public static void main(String[] args) throws IOException { if(args.length < 1) { System.err.println("Usage: java CisTransTest filename"); System.exit(0); } // create importer for the file argument String s = (String)args[0]; MolImporter molimp = new MolImporter(s); // store the imported molecules in m Molecule m = new Molecule(); // counter for molecules int n = 0; while(molimp.read(m)){ // read molecules from the file ++n; // increment counter System.err.println("mol "+n); // calculate double bond stereo for every double bond // which have at least one ligand on each node of the double bond for (int i=0; i<m.getBondCount(); i++){ MolBond b = m.getBond(i); if (b.getType() == 2){ // get the default frame CNode c1 = b.getCTAtom1(); CNode c2 = b.getNode1(); CNode c3 = b.getNode2(); CNode c4 = b.getCTAtom4(); if (c1 != null && c4 != null){ // cis/trans stereo for the default frame int ct = m.getStereo2(b, c1, c4, true); System.out.println(m.indexOf(c1)+"-"+ m.indexOf(c2)+"="+m.indexOf(c3)+"-"+ m.indexOf(c4)+" "+ ((ct == MolBond.CIS) ? "CIS" : (ct == MolBond.TRANS) ? "TRANS" : (ct == (MolBond.TRANS|MolBond.CIS)) ? "CIS|TRANS" : (""+ct) )+ (((ct & StereoConstants.CTUNSPEC) != 0) ? "CTUNSPEC" : " ") ); // E/Z stereo ct = m.getStereo2(b); System.out.println("E/Z "+ m.indexOf(c2)+"="+m.indexOf(c3)+" "+ ((ct == MolBond.CIS) ? "Z" : (ct == MolBond.TRANS) ? "E" : ""+ct )+ (((ct & StereoConstants.CTUNSPEC) != 0) ? "CTUNSPEC" : "") ); } } } } } }