Solubility Predictor
This manual gives you a walk-through on how to use the Solubility Predictor:
Introduction
Aqueous solubility is one of the most important physico-chemical properties in modern drug discovery. It has impact on ADME-related properties like drug uptake, distribution and even oral bioavailability. Solubility can also be a relevant descriptor for property-based computational screening methods in the drug discovery process. Hence there is a significant interest in fast, reliable, structure-based methods for predicting solubility in water for promising drug candidates.
Predicting solubility for drug molecules is not a trivial computational task. The used model has to somehow calculate the intrinsic solubility for the compound, which is a crucial parameter.
Intrinsic aqueous solubility is the equilibrium (thermodynamic) solubility of an ionizable compound at a pH where it is fully un-ionized (neutral).
ChemAxon's Solubility Predictor is able to calculate intrinsic solubility. The prediction uses a fragment-based method that identifies different structural fragments in the molecule and calculates their solubility contribution. The implementation is based on the article of Hou et al.
The figure below shows a molecule split up into fragments that are used in the intrinsic solubility prediction.
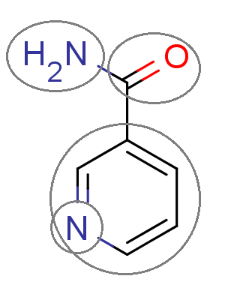
Fig. 1 The molecule is split into fragments to predict its solubility.
However, in many applications (such as therapeutic or lab measurements) it is also important to know the solubility in acidic/basic solutions at a given pH. Therefore the model should also be able to calculate the pH--solubility profile of a compound. The original solubility predictor method has been extended so that it is able to calculate the pH--dependent solubility.
The predictor can provide quantitative results, giving the solubility in logS, mg/mL or mol/L units. The predictive accuracy of the plugin is considered to be 1 logS unit. In case only an estimation about how well the compound is soluble, the plugin can give a solubility category as a qualitative measure.
On the logS unit
The logS is a common unit for measuring solubility. This unit is the 10-based logarithm of the solubility measured in mol/l unit, that is logS = log (solubility measured in mol/l).
The accuracy of the intrinsic solubility prediction was tested using two test sets in Hou et al. The two plots below show the experimental vs. predicted intrinsic solubility values and the deviation.
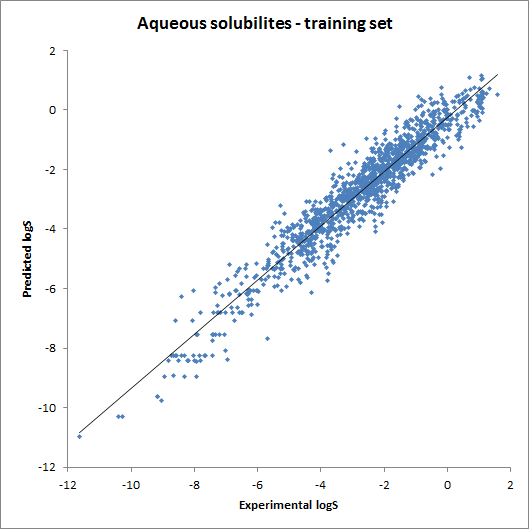
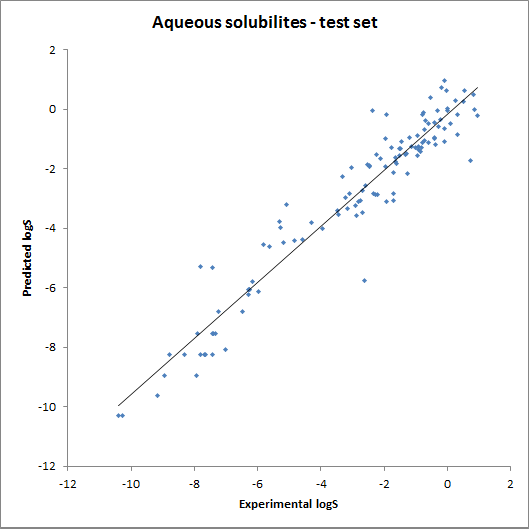
Fig. 2 Plots showing predicted vs. experimental intrinsic solubility values (see the reference 1. for data)
Tests for pH--logS profile were also run. The two plots below show calculated and experimental pH-logS profiles for different acidic, basic and zwitter-ionic compounds.
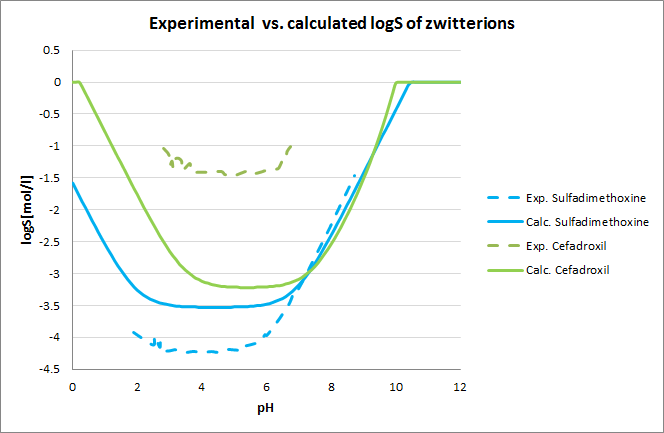
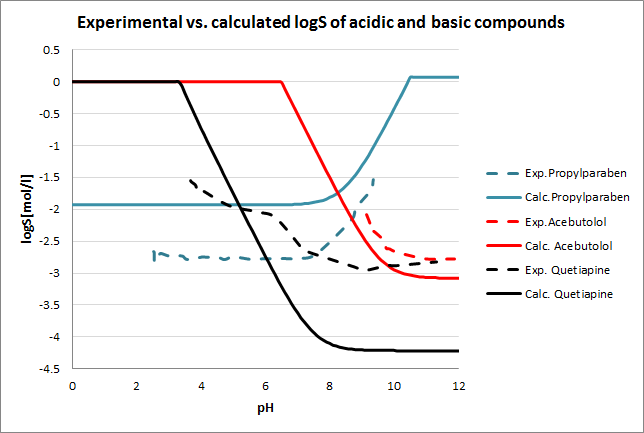
Fig. 3 Plots showing calculated vs. experimental pH-logS profiles for different compound sets (see the reference 2. for data)
Future goals
The Solubility Predictor will be developed further in the future. Among our future goals we have extending the prediction with a descriptor-based method and adding training features.
Usage
The Solubility Predictor is currently available in three ways.
MarvinSketch
The solubility predictor is integrated into Marvin Sketch as a plugin, which makes prediction fast and easy. The Solubility Plugin can be reached via the Calculations > Solubility > Aqueous Solubility menu item.
Menu system
The menu system of the Solubility Plugin has File, Options and Help menu items.
Result window
The result window displays the pH-logS plot and a table with prediction results given in one of the following ways :
-
Solubility information: displays intrinsic solubility value, solubility at pH 7.4 and a qualitative solubility category. These categories are:
-
low: if solubility is < 0.01 mg/ml
-
moderate: if solubility is between 0.01 and 0.06 mg/ml
-
high: if solubility is > 0.06 mg/ml
-
About qualitative categories
These categories are determined based on intrinsic solubility values.
-
pH-Solubility table: displays a table containing the predicted pH-solubility values.
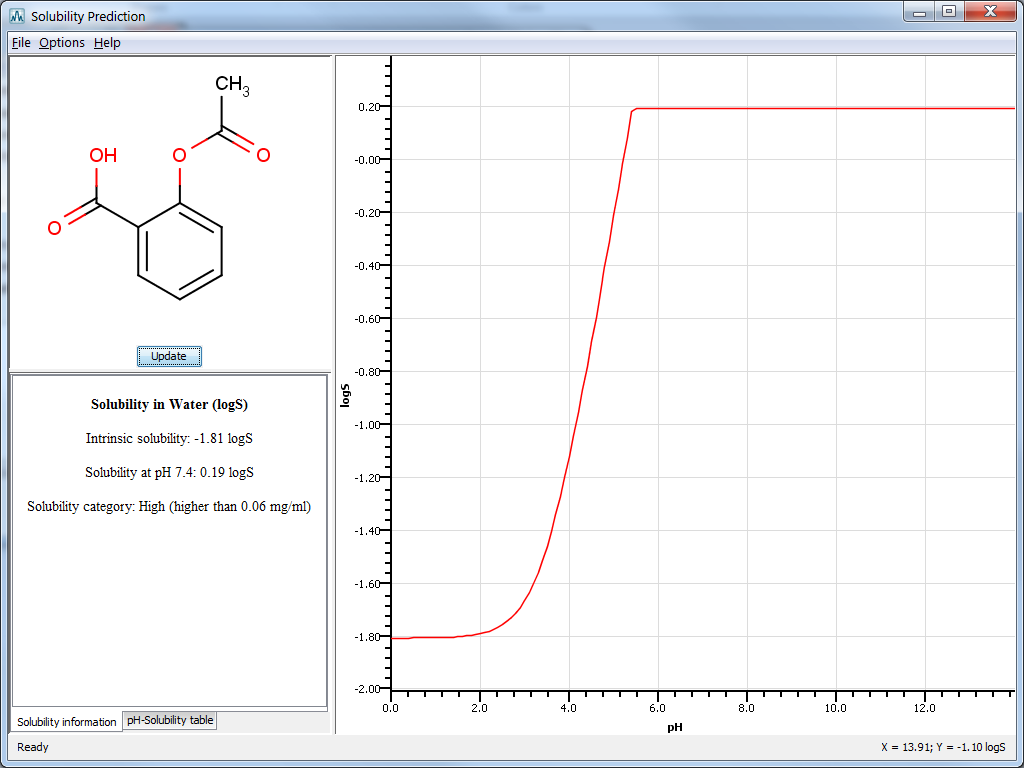
Fig. 4 The Solubility Plugin result window in MarvinSketch
cxcalc
The solubility predictor is integrated into the cxcalc command line tool. The command syntax is
cxcalc [general options] [input files/strings] logs [logs options] [input files/strings]
where the logS options are the following:
logs options: -h --help this help message -U --unit measurement unit [mg/ml | mol/l | logS] (default: logS) -i --intrinsic intrinsic solubility (default: false) -c --category solubility category (default: false) -H --pH solubility at this pH (default: not set) -l --lower (default: 0) -u --upper (default: 14) -s --step (default: 1)
Some examples on how to use the solubility predictor via cxcalc:
-
Calculating intrinsic solubility and solubility at pH 7.4 for compounds:
cxcalc logs -i true -H 7.4 molecules.smiles -
Calculating solubility values between pH 7.0 and 13.0 in mol/l unit:
cxcalc logs -U mol/l -l 7.0 -u 13.0 molecules.smiles -
Predicting qualitative solubility category:
cxcalc logs -c true test.mol
Chemical Terms
The Solubility Predictor is also integrated into ChemAxon's Chemical Terms language. The solubility can be calculated by the logS() function that has two parameters:
-
A unit parameter, which could be 'mg/ml', 'mol/l', 'logS', or 'category'. In case of 'category' is used, qualitative solubility is calculated. If no unit parameter is given, logS unit is used.
-
A pH parameter, which is a real number (e.g. '7.4') and sets the pH for solublity calculation. If no pH parameter is given, intrinsic solubility is calculated.
The following examples show how the functions above can be used with the Chemical Terms evaluator command line tool:
-
Calculating intrinsic solubility for acetic acid in logS unit:
evaluate -e "logS()" "CC(O)=O" -
Calculating solubility at pH 7.0 in mol/l unit:
evaluate -e "logS('7.0', 'mol/l')" molecule.mol -
Determining qualitative solubility category at pH 7.4:
evaluate -e "logS('7.4', 'category')" molecule.mol
KNIME
Solubility prediction is also available as a node in the KNIME workflow management system. All options that are available for solubility prediction are also available in KNIME. The pictures below show the General and Advanced Options of the node:
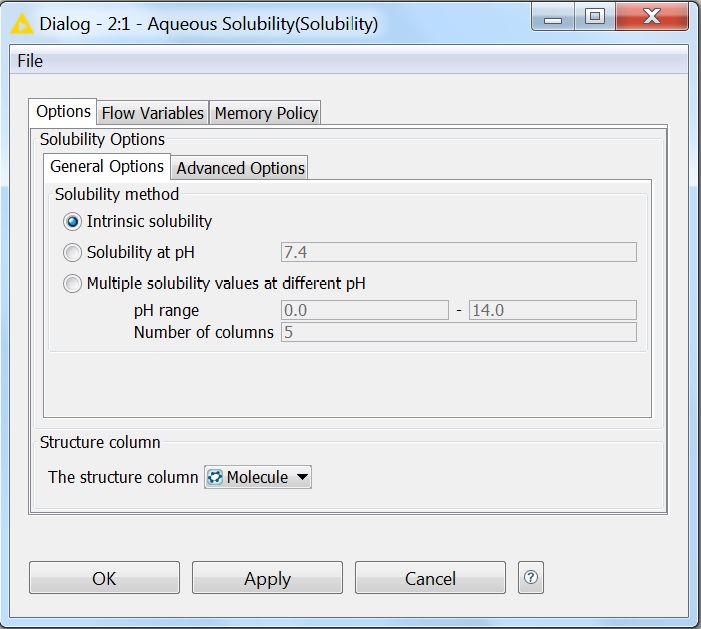
Fig. 5 General options of the Solubility KNIME node
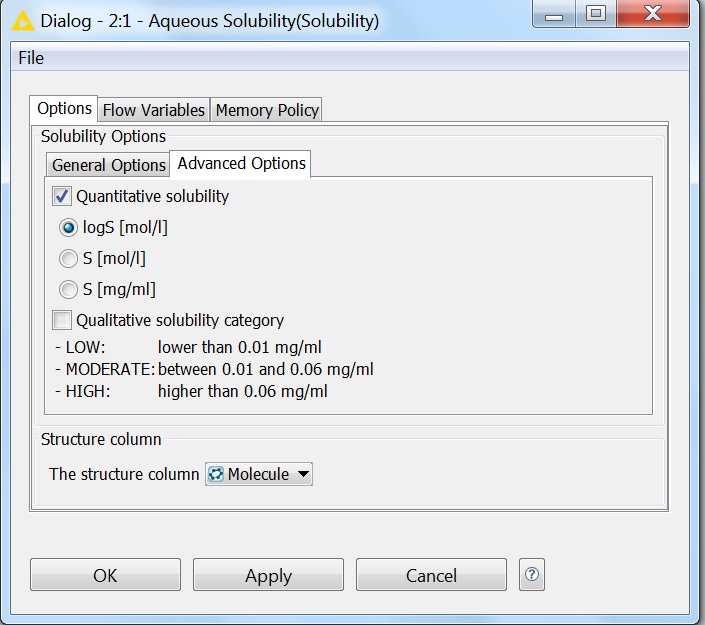
Fig. 6 Advanced options of the Solubility KNIME node
Example
The following simple KNIME workflow is an example of how to predict solubility using KNIME nodes:
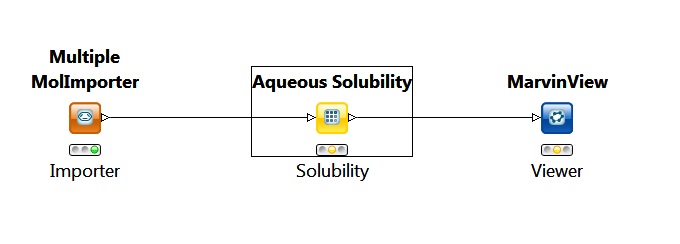
Fig. 7 Simple workflow for calculating solubility in KNIME
In this workflow the importer node provides the molecules for the solubility node. The solubility node is set to calculate the intrinsic qualitative and quantitative solubility for the input molecules.
The output in this case is a MarvinView node that is used to view the molecules with the calculated solubility values.
The picture above shows the state of the nodes before the running of the whole workflow.
The first few lines of the output viewed in the MarvinView node:
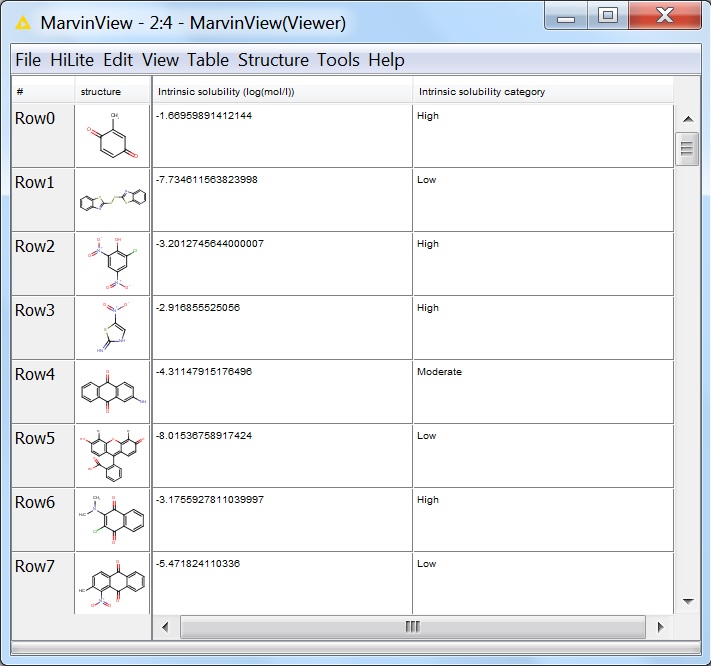
Fig. 8 First few lines of the output of the solubility calculation
References
1. Hou, T. J.; Xia, K.; Zhang, W.; Xu, X. J. ADME Evaluation in Drug Discovery. 4. Prediction of Aqueous Solubility Based on Atom Contribuition Approach. J. Chem. Inf. Comput. Sci. 2004, 44, 266-275
2. Shoghi, E.; Fuguet, E.; Bosch, E.; Rafols, C. Solubility-pH profiles of some acidic, basic and amphoteric drugs European Journal of Pharmaceutical Sciences 2013, 48, 291-300
 Export to PDF: exports the results into a PDF file, which can be used as a report.
Export to PDF: exports the results into a PDF file, which can be used as a report.