Strip Salts
This action removes predefined fragments from multifragment molecules (regarded as salts), and saves the information of the removed fragments in SDF property fields. The salts will be removed only in case of exact isotope match (see the examples). A built-in salt dictionary is used by default, but custom salt definitions can be used by setting it in place during action configuration.
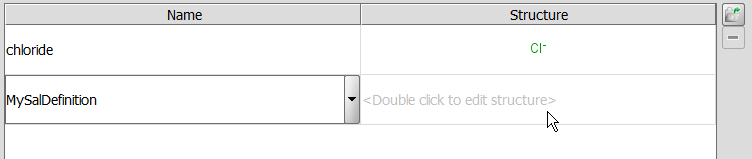
Custom salt definition: You can edit your custom list of salt definitions after you placed action "Strip Salt" to the right side panel of "Configure Actions" window. Select the action on the right side panel and edit the salt definitions on the lower panel.
Strip Salts Editor
Below column "Name", you can type a desired salt name or select one from the drop-down list.
Edit the appropriate structure by double clicking on the field below column "Structure" (a MarvinSketch window will appear).
You can delete any table element as follows: Click on the relevant structure field; the row will be highlighted. Click on  .
.
Built-in salt dictionary: You can load the elements of built-in salt dictionary by clicking on button  .
.
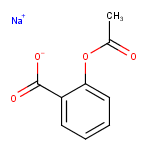
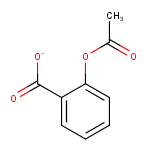
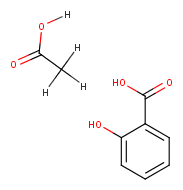
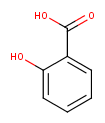
Example :
For the Strip Salts action Sodium ion (Na+) and Acetic acid were listed as salts to be stripped.
|
Action |
Input |
Output |
|
Strip Salts |
|
|
|
Strip Salts |
|
|
|
- |
|
|
If the salt is not found to be an exact isotope match, it is not removed, the Strip Salts action will not run.
Option: Don't remove last component
This Standardizer action does not remove all components (fragments) from the molecule by default. In this case, if all components of the molecule are defined in the salt dictionary then the largest (based on molecular weight) is not removed. If you clear the checkbox, all fragments that are present in the salt list will be removed.
The removed salts are stored in the "Salt<n>" property.