Generate3D
This page demonstrates the usage and the theory behind Generate3D:
Introduction
Generate3D is ChemAxon's module for 3D coordinate/conformer generation and analysis.
Where is Generate3D available ?
MarvinSketch
Generate3D can be reached as Clean3D under Structure > Clean3D on the MarvinSketch GUI. You can draw or import your molecule directly onto the canvas, and Clean3D generates 3D coordinates for the structure. Ctrl + 3 serves as a shortcut for the same functionality on the canvas. The generated 3D structure can be rotated on the canvas after pressing the F7 shortcut.
To get more information on the usage of Clean3D in Marvin, see the corresponding part of the user manual.
Five cleaning methods are supported in Marvin; each represents a different chemical use case scenario:
-
Fine Build: this generates 3D coordinates taking into account the implicit hydrogens of the initial stucture; the hydrogens however are not presented in the final 3D structure.
-
Fine with Hydrogenize: the same as the Fine Build, but it also returns explicit hydrogens in the final 3D structure.
-
Fast Build: this build uses a different, but in some cases faster method to generate coordinates than Fine Build. The justification of this method is the need for a rough 3D structure that is not necessarily valid chemically, but can be built fast. If fast clean fails, fine clean is used instead, and any generated structure is returned.
-
Build or Optimize: this build generates 3D coordinates for non-3D structures, otherwise it runs Dreiding a geometry optimization based on the Dreiding force-field starting from the initial structure.
-
Optimize: this only runs a Dreiding force field based geometry optimization starting from the initial stucture.
These methods are available as options under Structure > Clean3D > Cleaning Method.
Molconvert
The functionality of Generate3D is also available in the molconvert command line tool. This enables the user to sequentially generate 3D coordinates for many initial structures.
Use the
molconvert -3:options inputfilecommand to batch-process your inputfile. To get a full list of options you can set within molconvert, use the
molconvert -H3D command.
Some 3D coordinate generation examples:
-
Cleaning a molecule given as a SMILES string, and exporting the 3D structure to MOL:
molconvert -3:c3 mol -s "C1CC3C(C1)C4C2CC2CC34" -o struct3d.molThis gives the same result as the option S{fine}.
-
Cleaning a library of molecules stored in SDF, skipping the ones that already have 3D coordinates:
molconvert -3:c31 mol library.sdf -o cleanedlib.mol -
Generating fine 3D geometry for a molecule given as a SMILES string, using MMFF94 force field optimization, with strict convergence criterion on the gradient:
molconvert -3:S{fine}[mmff94]L{3} mol -s "CC(CCC)CCCCl" -o struct3d.mol -
Generating 3D coordinates for a library of molecules in SDF, using the fine method and a time limit of 6000 seconds pre-set by the user. We also want to store the MMFF94 energies for each molecule as an SDF property:
molconvert -3:S{fine}[mmff94][timelimit]{6000}[E] mol library.sdf -o cleanedlib.sdf -
Generating 3D coordinates for a molecule given as a SMILES string, while adding explicit hydrogens and using very strict optimization criterion:
molconvert -3:S{fine}[prehydrogenize][o]{1}{3} mol -s "CCCCC(CCC)C" -o struct3d.mol
MarvinBeans API
The Generate3D component can also be used via the Marvin Beans API. The options of molconvert can also be set through the API, using the setClean3dOptions method.
Chemical Terms
One can also use ChemAxon's Chemical Terms to generate 3D structures in the following ways:
-
In JChem for Excel the 3D cleaning functionality is available through the JC3DCleanStructure function. For more information on this feature, see the corresponding part of the JChem for Excel manual.
-
The molconvert Chemical Terms function is capable of generating 3D coordinates for the initial structure, and returning it in a given format string. See the description of the function in the the manual.
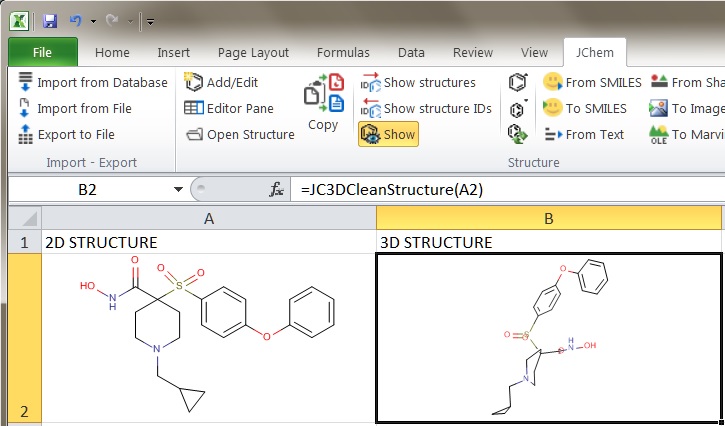
Fig. 1 Generating 3D coordinates in JChem for Excel using the JC3DCleanStructure function
Standardizer
In Standardizer the 3D cleaning functionality is available through the clean:3 action. For more information on this feature, see the corresponding part of the Standardizer manual.
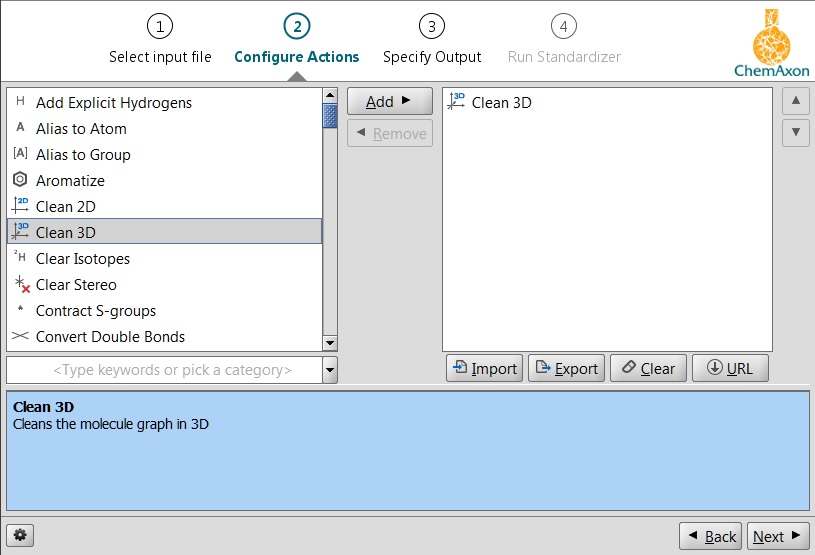
Fig. 2 Standardizer clean3D function
Conformer generation and analysis
Generate3D is also used for generating conformers for a given initial structure. Conformer generation is available in three different ways.
Conformer Plugin
Conformers Plugin is a calculator plugin of ChemAxon that offers the conformer generation functionality in MarvinSketch. The plugin can be reached under Calculations > Conformation > Conformers.
The documentation of the plugin, with all of its options, can be found here .
cxcalc
The conformer generation and analysis functionality of the conformer plugin can also be reached via ChemAxon's command line calculator cxcalc . The same options can be used as with molconvert. For details see the corresponding part of the manual.
molconvert
Molconvert offers a fully customizable way to generate conformers. The conformer generation and analysis functionality of Generate3D can be invoked using the -3:[ca]{v1}{v2} option. The two parameter values v1, v2 sets the number of generated and stored conformers, respectively. The conformers are stored as molecular properties.
Some conformer generation examples with molconvert :
-
Without any count parameters, a warning is given, and only one conformer is generated and reported:
molconvert -3:[ca] -s "CCC(CC)CCCCl" -o conf3d.sdf -
Setting the number of the generated and stored conformers to 10:
molconvert -3:[ca]{10} sdf -s "CCC(CC)CCCCl" -o conf3d.sdf -
Generating at least 5 conformers, reporting at most 10 of them:
Note that if the number of reported conformers is smaller than that of the generated conformers, a warning is given, and the former is set to the latter.
molconvert -3:[ca]{5}{10} sdf -s "CCC(CC)CCCCl" -o conf3d.sdf -
Invoking sophisticated low-energy conformer prediction, using MMFF94 force field:
molconvert -3:[hyperfine][mmff94] sdf -s "CCC(CC)CCCCl" -o conf3d.sdf -
Generating and reporting conformers with customized conformer RMSD diversity criterion 0.05:
molconvert -3:[ca]{5}{20}[diversity]{0.05} sdf -s "CCC(CC)CC" -o conf3d.sdf
Instant JChem
The conformer generation and analysis functionality can also be reached via the Chemical Terms conformation evaluation functions. A list of these functions and some examples of their usage are available in the corresponding part of the Chemical Terms manual.
ChemAxon workflow tools
Workflow management systems were designed to easily compose and execute complex computational tasks, such as data mining, data processing, or statistical analysis. In order to handle the more specific needs of chemistry informatics within this clean and straightforward framework, ChemAxon technology has been implemented into three major scientific workflow management systems: KNIME, Pipeline Pilot, and Inforsense.
KNIME
JChem Extensions offers a set of KNIME nodes with which the user can easily build their own workflows for handling chemical data. KNIME also enables users to integrate their own softwares and other tools. Among JChem Extensions there are nodes that can perform 3D coordinate generation, conformer generation and analysis. To have more information on building chemical workflows using KNIME, see the
ChemAxon KNIME workflow manual
.
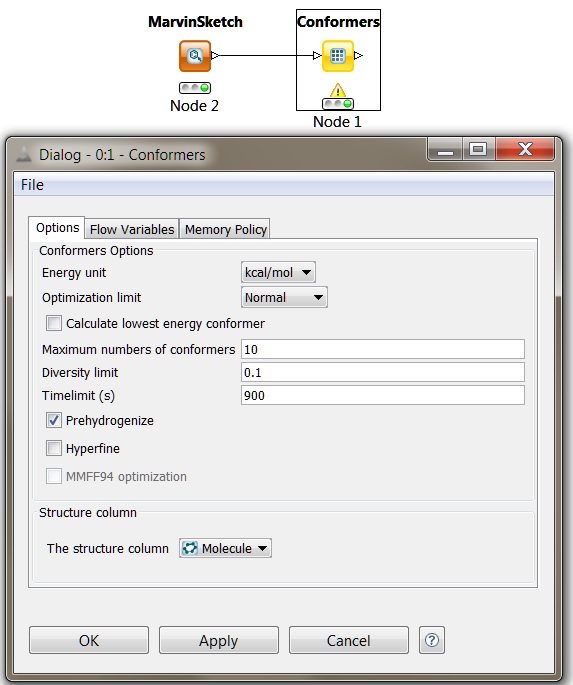
Fig. 3 KNIME conformer generation node
Pipeline Pilot
Currently, about 16 different ChemAxon tools are available in Pipeline Pilot, another chemical informatics platform. Among these there are components also for 3D coordinate generation, conformer generation and analysis. To have more informations on building chemical workflows using Pipeline Pilot, see the ChemAxon Pipeline Pilot workflow manual .
Theory behind coordinate/conformer generation with Generate3D
Structure building process
The algorithm behind 3D coordinate generation uses a divide-and-conquer approach. First, the initial structure is split into small fragments. Then the fragments are organized into a build tree using their original connectivity information. The tree is built in a way to represent the connectivity of the fragments before the split. Each node stores a fragment: the leaves of the tree store the small fragments, while non-leaf nodes store fragments obtained from the fusion of smaller ones at lower levels. The coordinate and conformer generation is done in a command-driven fashion. Every node can be instructed to generate 3D coordinates or conformers for the represented fragment. Leaves do this in an atom-by-atom manner, determining energetically favourable, multiple-atom displacements. Other nodes - including the root - proceeds by exploring the conformational space of fused conformers that were generated by the children of the node. If no more conformers can be generated at a given node, a new conformer from a child is returned, and the fusion process restarts.
Geometry optimization
Conformers from the fusion process are optimized when required before passing to a higher level node in the build tree. This happens when the rigid fuse results in high energy conformation (due to e.g. strained bond/angle, atom proximity or clash). Conformers generated for the initial structure (represented by the root node in the build tree) are also optimized. The building process uses a proprietary extended version of the Dreiding force field. Optionally an additional MMFF94 based optimization and energy calculation can be run on these final structures.
Conformer generation options
Diversity for the conformer generation can be pre-set using the diversity limit option. Two conformers having greater root-mean-square-deviation (RMSD) than the limit are considered different. Generated conformers can reside in invalid local minimum on the potential energy surface after geometry optimization. This can be eliminated by a hyperfine post-processing step: every conformer is passed through short, low-temperature molecular dynamic runs, followed by a strict geometry optimization.
For details on the algorithm behind Generate3D, see the references below.
References
-
ChemAxon article: Molecules from the Minkowski space: an approach to building 3D molecular structures
-
ChemAxon poster: 3D structure prediction and conformational analysis
-
ChemAxon poster: Advanced automatic generation of 3D molecular structures