Cis Trans stereoisomers in 0 Dimension
If the atomic coordinates are all zero then the molecule's dimension is zero. The double bond stereo information is stored in the bond flag referring to the first ligand (which is not the other end of the double bond) of each endpoint for the double bond (default reference frame).
Setting cis/trans information in 0D
The cis/trans information can be set directly using the setFlags(int) function of the MolBond class. In this case the set value refers to the default reference frame arrangement a1-a2=a3-a4 where a2 and a3 is the double bond first and second node and a1 and a4 is the first ligand of a2 and a3 respectively.
Code example for getting the default reference frame:
MolBond b = molecule.getBond(2) // default reference frame // for which the bond stereo will be set MolAtom a1 = b.getCTAtom1(); MolAtom a2 = b.getAtom1(); MolAtom a3 = b.getAtom2(); MolAtom a4 = b.getCTAtom4();
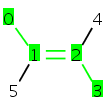
Code example to set flags using the default reference frame:
MolBond b = molecule.getBond(2); // set CIS value to the reference frame b.setFlags(StereoConstants.CIS, StereoConstants.CTUMASK);
Another possibility to set cis/trans information is via the setStereo2Flags(MolAtom a1, MolAtom a4, int s) function of the MolBond class, where the reference frame is defined by the a1 and a4 atoms.
Code example using given reference frame:
MolBond b = molecule.getBond(2); // reference frameMolAtom a1 = molecule.getAtom(0); MolAtom a4 = molecule.getAtom(4); // set CIS value to the reference frameb.setStereo2Flags(a1, a4, StereoConstants.TRANS);
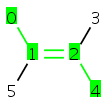
Getting cis/trans information in 0D
The cis/trans information can be got directly by the getFlags() function of the MolBond class. In this case the value returned refers to the default reference frame.
Code example:
MolBond b = molecule.getBond(2); int s = b.getFlags() & StereoConstants.CTUMASK; 
s = TRANS
With a given reference frame there are four options to get the stereo information:
-
1. getStereo2(MolAtom a1, int i2, int i3, MolAtom a4)
-
2. getStereo2(int i1, int i2, int i3, int i4)
-
3. getStereo2(MolBond b, MolAtom a1, MolAtom a4)
-
4. getStereo2(MolBond b, MolAtom a1, MolAtom a4, boolean grcheck)
These all functions of the MoleculeGraph class. The difference between them is how the reference frame is given (by the node or by the node index) and the last method can check atom equivalences too.
Code example:
MolBond b = molecule.getBond(2); // reference frame MolAtom a1 = molecule.getAtom(0); MolAtom a4 = molecule.getAtom(4); int s = molecule.getStereo2(b, a1, a4); 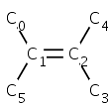
s = CIS
Code example with atom equivalence check:
MolBond b = molecule.getBond(2);// reference frameMolAtom a1 = molecule.getAtom(0);MolAtom a4 = molecule.getAtom(4);int s = molecule.getStereo2(b, a1, a4, true); 
s = 0